Genetic Determinants of Immune Cells and Hepatocellular Carcinoma Risk: A Bioinformatics and Bidirectional Mendelian Randomization Study
-
摘要:目的
基于生物信息学及特定算法筛选肝细胞癌的核心靶点并探讨其与免疫细胞的关系,并通过孟德尔随机化方法探讨免疫细胞与肝细胞癌的因果关系。
方法通过GEO和TCGA数据库对肝细胞癌发生的相关基因进行筛选,并通过GSVA和CIBERSORT算法进行免疫浸润分析,随后对免疫细胞与肝细胞癌的因果关系进行双向孟德尔随机化分析。
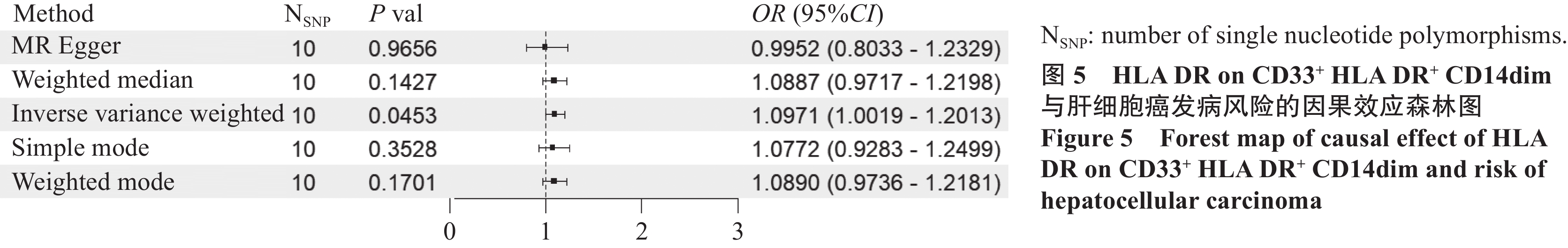
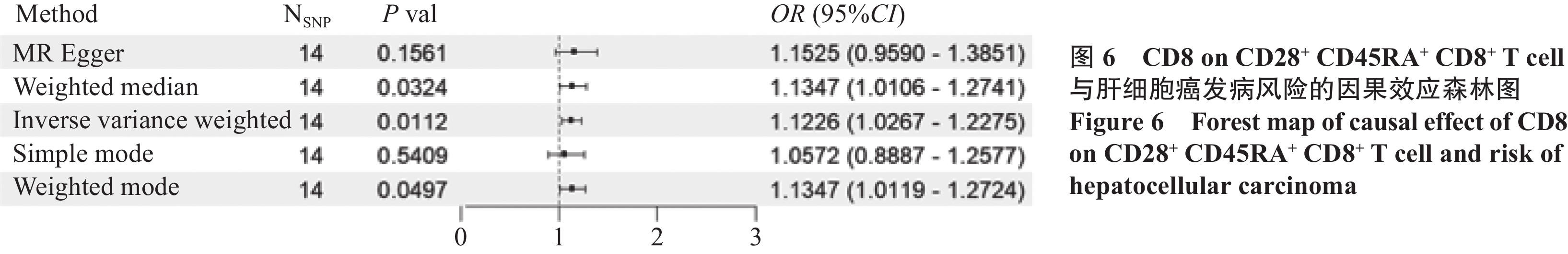
结果筛选出284个肝癌相关基因,在蛋白互作网络中获取到120个相关基因。孟德尔随机化结果显示:髓系细胞中的HLA DR on CD33+ HLA DR+ CD14dim(OR=1.097,95%CI: 1.002~1.201,P=0.045,PBonferroni=0.091)和调节性T细胞中的CD8 on CD28+ CD45RA+ CD8+ T cell(OR=1.123,95%CI: 1.027~1.228,P=0.011,PBonferroni=0.022)是肝细胞癌的危险因素;肝细胞癌是经典树突状细胞中的HLA DR++ monocyte Absolute Count(OR=0.812,95%CI: 0.702~0.938,P=0.005,PBonferroni=0.139)的保护因素。免疫浸润分析显示,关键基因与交集免疫细胞之间具有较好的相关性。
结论肝癌的发生发展可能与CDK1、CCNB1、CDC20有关,并与Th2 cells、T helper cells及Th17 cells、DC等呈现较高程度的相关性,孟德尔随机化显示HLA DR on CD33+、HLA DR+ CD14dim和CD8 on CD28+、CD45RA+ CD8+ T cell与肝细胞癌的风险增加有关,而肝细胞癌的发生风险与HLA DR++ monocyte Absolute Count的水平降低有关。
Abstract:ObjectiveTo identify core targets of hepatocellular carcinoma (HCC) by using bioinformatics and specific algorithms, explore their relationships with immune cells, and investigate the causal relationships between immune cells and HCC through Mendelian randomization.
MethodsRelevant genes associated with the development of HCC were screened using the GEO and TCGA databases. Immune infiltration analysis was conducted using GSVA and CIBERSORT algorithms. A bidirectional Mendelian randomization analysis was then performed to explore the causal relationships between immune cells and HCC.
ResultsA total of 284 HCC-related genes were identified, with 120 genes recognized within the protein interaction network. Immune infiltration analysis revealed significant correlations between key genes and immune cells. Mendelian randomization results indicated that HLA DR on CD33+ HLA DR+ CD14dim (OR=1.097, 95%CI: 1.002–1.201, P=0.045, PBonferroni=0.091) and CD8 on CD28+ CD45RA+ CD8+ T cell (OR=1.123, 95%CI: 1.027–1.228, P=0.011, PBonferroni=0.022) were the risk factors for HCC. Conversely, HLA DR++ monocyte absolute count was identified as a protective factor for HCC (OR=0.812, 95%CI: 0.702–0.938, P=0.005, PBonferroni=0.139).
ConclusionThe occurrence and development of liver cancer may be related to CDK1, CCNB1, and CDC20, showing a high degree of correlation with Th2 cells, T helper cells, Th17 cells, and DCs. Mendelian randomization shows that HLA DR on CD33+HLA DR+ CD14dim and CD8 on CD28+CD45RA+CD8+T cells are associated with an increased risk of HCC. The risk of hepatocellular carcinoma is associated with a decrease in the level of HLA DR++monocyte absolute count.
-
Key words:
- Mendelian randomization /
- Hepatocellular carcinoma /
- Immune cell /
- Immunophenotype /
- SNPs
-
Competing interests: The authors declare that they have no competing interests.利益冲突声明:所有作者均声明不存在利益冲突。作者贡献:吴 桐:研究方法和数据分析、论文撰写高 菲:研究方法的完善和修改、数据分析滕 飞:数据检索和下载、数据分析张巧丽:论文审阅和修改
-
表 1 正向MR敏感性分析
Table 1 Positive MR sensitivity analysis
Panel Trait IVW MR-Egger Q P Intercept P Blood protein measurement HLA DR on CD33+ HLA DR+ CD14dim 12.0333 0.3611 0.0305 0.3822 Blood protein measurement CD8 on CD28+ CD45RA+ CD8+ T cell 23.9265 0.0911 − 0.0059 0.8276 表 2 反向MR敏感性分析
Table 2 Reverse MR sensitivity analysis
Panel Trait IVW MR-Egger Q P Intercept P Lymphocyte count CD20- CD38- B cell %B cell 0.2154 0.8979 0.0304 0.7262 Leukocyte count HLA DR++ monocyte %leukocyte 1.0918 0.5793 − 0.0504 0.6030 Leukocyte count HLA DR++ monocyte absolute count 1.8067 0.4052 − 0.0773 0.4596 Myeloid white cell count Monocytic myeloid-derived suppressor cells absolute count 0.8472 0.6547 0.0712 0.5785 Lymphocyte count Transitional B cell %lymphocyte 1.2605 0.5325 − 0.0634 0.5193 Blood protein measurement BAFF-R on IgD+ CD38+ B cell 1.3778 0.5021 − 0.0299 0.7616 Blood protein measurement BAFF-R on transitional B cell 0.1478 0.9288 − 0.0059 0.9462 Blood protein measurement CD24 on IgD+ CD38+ B cell 0.7720 0.6798 − 0.0293 0.7390 Blood protein measurement CD27 on IgD+ CD38- unswitched memory B cell 0.4226 0.8095 0.0097 0.9333 Blood protein measurement CD38 on CD20- B cell 2.2254 0.3287 0.0506 0.6504 Blood protein measurement IgD on IgD+ CD38- B cell 2.2311 0.3277 0.0958 0.3954 Blood protein measurement CD3 on Effector Memory CD8+ T cell 0.4769 0.7878 0.0160 0.8661 Blood protein measurement CD3 on HLA DR+ CD4+ T cell 0.3367 0.8450 − 0.0239 0.7976 Blood protein measurement CD86 on granulocyte 1.2054 0.5473 0.0699 0.5245 Blood protein measurement CD33 on CD33+ HLA DR+ CD14dim 0.8884 0.6413 0.0914 0.5288 Blood protein measurement CD33 on Monocytic Myeloid-Derived Suppressor Cells 1.5625 0.4578 0.0829 0.5587 Blood protein measurement CD33 on CD33dim HLA DR- 0.5776 0.7492 0.0729 0.5994 Blood protein measurement CD33 on basophil 0.5990 0.7412 0.0696 0.6138 Blood protein measurement CD33 on CD33+ HLA DR+ 0.9939 0.6084 0.0969 0.5098 Blood protein measurement CD33 on CD33+ HLA DR+ CD14- 1.0684 0.5861 0.0990 0.5034 Blood protein measurement CD4 on HLA DR+ CD4+ T cell 0.2144 0.8984 0.0294 0.7503 Blood protein measurement FSC-A on monocyte 1.5506 0.4606 0.0926 0.4311 Blood protein measurement CD64 on CD14- CD16+ monocyte 0.6259 0.7313 0.0311 0.7254 Blood protein measurement CCR2 on CD14+ CD16+ monocyte 0.2206 0.8956 0.0304 0.7223 Blood protein measurement CD4 on Effector Memory CD4+ T cell 0.2212 0.8953 0.0107 0.9076 Blood protein measurement CD8 on Effector Memory CD8+ T cell 1.1106 0.5739 − 0.0772 0.4850 Blood protein measurement CD8 on Terminally Differentiated CD8+ T cell 3.0539 0.2172 0.0729 0.6119 Blood protein measurement SSC-A on myeloid Dendritic Cell 1.5909 0.4514 0.0678 0.5353 Blood protein measurement SSC-A on monocyte 1.4362 0.4877 0.0904 0.4439 -
[1] Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-263. doi: 10.3322/caac.21834
[2] Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2021, 7(1): 6.
[3] Thomas London W, Petrick JL, et al. Liver cancer. In: M Thun, MS Linet, JR Cerhan, CA Haiman, D Schottenfeld, eds. Cancer Epidemiology and Prevention[M]. 4th ed. Oxford University Press, 2018: 635-660.
[4] 中华人民共和国国家卫生健康委员会. 原发性肝癌诊疗指南(2024年版)[J]. 临床肝胆病杂志, 2024, 40(5): 893-918. [National Health Commission of the People's Republic of China. Primary liver cancer Diagnosis and treatment guidelines (2024 edition)[J]. Lin Chuang Gan Dan Bing Za Zhi, 2024, 40(5): 893-918.] doi: 10.12449/JCH240508 National Health Commission of the People's Republic of China. Primary liver cancer Diagnosis and treatment guidelines (2024 edition)[J]. Lin Chuang Gan Dan Bing Za Zhi, 2024, 40(5): 893-918. doi: 10.12449/JCH240508
[5] Massarweh NN, El-Serag HB. Epidemiology of hepatocellular carcinoma and intrahepatic cholangiocarcinoma[J]. Cancer Control, 2017, 24(3): 1073274817729245.
[6] 刘宗超, 李哲轩, 张阳, 等. 2020全球癌症统计报告解读[J]. 肿瘤综合治疗电子杂志, 2021, 7(2): 1-14. [Liu ZC, Li ZX, Zhang Y, et al. Interpretation of 2020 global cancer statistics report[J]. Zhong Liu Zong He Zhi Liao Dian Zi Za Zhi, 2019, 7(2): 1-14.] Liu ZC, Li ZX, Zhang Y, et al. Interpretation of 2020 global cancer statistics report[J]. Zhong Liu Zong He Zhi Liao Dian Zi Za Zhi, 2019, 7(2): 1-14.
[7] de Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth[J]. Cancer Cell, 2023, 41(3): 374-403. doi: 10.1016/j.ccell.2023.02.016
[8] Yasuoka H, Asai A, Ohama H, et al. Increased both PD-L1 and PD-L2 expressions on monocytes of patients with hepatocellular carcinoma was associated with a poor prognosis[J]. Sci Rep, 2020, 10(1): 10377. doi: 10.1038/s41598-020-67497-2
[9] Cao D, Chen MK, Zhang QF, et al. Identification of immunological subtypes of hepatocellular carcinoma with expression profiling of immune-modulating genes[J]. Aging (Albany NY), 2020, 12(12): 12187-12205.
[10] Liu F, Qin L, Liao Z, et al. Microenvironment characterization and multi-omics signatures related to prognosis and immunotherapy response of hepatocellular carcinoma[J]. Exp Hematol Oncol., 2020, 9: 10. doi: 10.1186/s40164-020-00165-3
[11] 黄燕妮, 蓝雪灵, 朱敏敏, 等. PD-1/PD-L1抑制剂联合抗血管内皮生长因子药物免疫治疗晚期肝癌的研究进展[J]. 中国药理学通报, 2024, (8): 1429-1436. [Huang Yanni, LAN Xueling, Zhu Minmin, et al. Research progress of PD-1/PD-L1 inhibitors combined with anti-vascular endothelial growth factor drugs in immunotherapy of advanced liver cancer[J]. Zhongguo Yao Li Xue Tong Bao, 2024, (8): 1429-1436.] Huang Yanni, LAN Xueling, Zhu Minmin, et al. Research progress of PD-1/PD-L1 inhibitors combined with anti-vascular endothelial growth factor drugs in immunotherapy of advanced liver cancer[J]. Zhongguo Yao Li Xue Tong Bao, 2024, (8): 1429-1436.
[12] Luo X, Huang W, Li S, et al. SOX12 Facilitates Hepatocellular Carcinoma Progression and Metastasis through Promoting Regulatory T-cells Infiltration and Immunosuppression[J]. Adv Sci (Weinh), 2024, 11(36): e2310304.
[13] Haycock PC, Burgess S, Wade KH, et al. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies[J]. Am J Clin Nutr, 2016, 103(4): 965-978. doi: 10.3945/ajcn.115.118216
[14] Sanderson E, Glymour MM, Holmes MV, et al. Mendelian randomization[J]. Nat Rev Methods Primers, 2022, 2: 6. doi: 10.1038/s43586-021-00092-5
[15] Orrù V, Steri M, Sidore C, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy[J]. Nat Genet, 2020, 52(10): 1036-1045. doi: 10.1038/s41588-020-0684-4
[16] Birney E. Mendelian Randomization[J]. Cold Spring Harb Perspect Med, 2022, 12(4): a041302.
[17] Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants[J]. Int J Epidemiol, 2011, 40(3): 740-752. doi: 10.1093/ije/dyq151
[18] Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations[J]. Bioinformatics, 2019, 35(22): 4851-4853. doi: 10.1093/bioinformatics/btz469
[19] Burgess S, Thompson SG. Interpreting findings from mendelian randomization using the MR-Egger method[J]. Eur J Epidemiol, 2017, 32(5): 377-389. doi: 10.1007/s10654-017-0255-x
[20] Wu F, Huang Y, Hu J, et al. Mendelian randomization study of inflammatory bowel disease and bone mineral density[J]. BMC Med, 2020, 18(1): 312. doi: 10.1186/s12916-020-01778-5
[21] Hao X, Ren C, Zhou H, et al. Association between circulating immune cells and the risk of prostate cancer: a Mendelian randomization study[J]. Front Endocrinol (Lausanne), 2024, 15: 1358416.
[22] Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015[J]. CA Cancer J Clin, 2016, 66(2): 115-132. doi: 10.3322/caac.21338
[23] Liao S, Wang K, Zhang L, et al. PRC1 and RACGAP1 are Diagnostic Biomarkers of Early HCC and PRC1 Drives Self-Renewal of Liver Cancer Stem Cells[J]. Front Cell Dev Biol, 2022, 10: 864051. doi: 10.3389/fcell.2022.864051
[24] Zongyi Y, Xiaowu L. Immunotherapy for hepatocellular carcinoma[J]. Cancer Lett, 2020, 470: 8-17. doi: 10.1016/j.canlet.2019.12.002
[25] Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases[J]. Trends Biochem Sci, 2005, 30(11): 630-641.
[26] Zou Y, Ruan S, Jin L, et al. CDK1, CCNB1, and CCNB2 are Prognostic Biomarkers and Correlated with Immune Infiltration in Hepatocellular Carcinoma[J]. Med Sci Monit, 2020, 26: e925289.
[27] Wang Z, Wan L, Zhong J, et al. Cdc20: a potential novel therapeutic target for cancer treatment[J]. Curr Pharm Des, 2013, 19(18): 3210-3214.
[28] Wang J, Amin A, Cheung MH, et al. Targeted inhibition of the expression of both MCM5 and MCM7 by miRNA-214 impedes DNA replication and tumorigenesis in hepatocellular carcinoma cells[J]. Cancer Lett, 2022, 539: 215677. doi: 10.1016/j.canlet.2022.215677
[29] Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation[J]. Science, 1996, 272(5258): 54-60. doi: 10.1126/science.272.5258.54
[30] Hamann D, Baars PA, Rep MH, et al. Phenotypic and functional separation of memory and effector human CD8 T cells[J]. J Exp Med, 1997, 186(9): 1407-1418. doi: 10.1084/jem.186.9.1407
[31] Pilch H, Hoehn H, Schmidt M, et al. CD8+CD45RA+CD27-CD28-T-cell subset in PBL of cervical cancer patients representing CD8+T-cells being able to recognize cervical cancer associated antigens provided by HPV 16 E7[J]. Zentralbl Gynakol, 2002, 124(8-9): 406-412.
[32] Höhn H, Jülch M, Pilch H, et al. Definition of the HLA-A2 restricted peptides recognized by human CD8+ effector T cells by flow-assisted sorting of the CD8+ CD45RA+ CD28- T cell subpopulation[J]. Clin Exp Immunol, 2003, 131(1): 102-110. doi: 10.1046/j.1365-2249.2003.02036.x
[33] Wu Z, Shi H, Zhang L, et al. Comparative analysis of monocyte-derived dendritic cell phenotype and T cell stimulatory function in patients with acute-on-chronic liver failure with different clinical parameters[J]. Front Immunol, 2023, 14: 1290445. doi: 10.3389/fimmu.2023.1290445
[34] Mengos AE, Gastineau DA, Gustafson MP. The CD14+HLA-DRlo/neg Monocyte: An Immunosuppressive Phenotype That Restrains Responses to Cancer Immunotherapy[J]. Front Immunol, 2019, 10: 1147. doi: 10.3389/fimmu.2019.01147
[35] Gustafson MP, Lin Y, Bleeker JS, et al. Intratumoral CD14+ Cells and Circulating CD14+HLA-DRlo/neg Monocytes Correlate with Decreased Survival in Patients with Clear Cell Renal Cell Carcinoma[J]. Clin Cancer Res, 2015, 21(18): 4224-4233.




 下载:
下载: