Exploration and Verification of Prognostic Value of Endothelial Cells in Glioblastoma
-
摘要:目的
探讨并验证胶质母细胞瘤中内皮细胞的预后价值。
方法通过对TCGA和CGGA数据库进行生物信息分析,按照一系列标准筛选GBM单细胞数据中内皮细胞相关标志物并进行单因素Cox回归分析,获取并筛选内皮细胞预后相关标志物,构建内皮细胞相关预后风险评分。qPCR实验验证预后相关标志物在GBM组织及瘤周正常脑组织中的表达差异。Kaplan-Meier法构建生存曲线以鉴定该预后风险评分的预后效能。
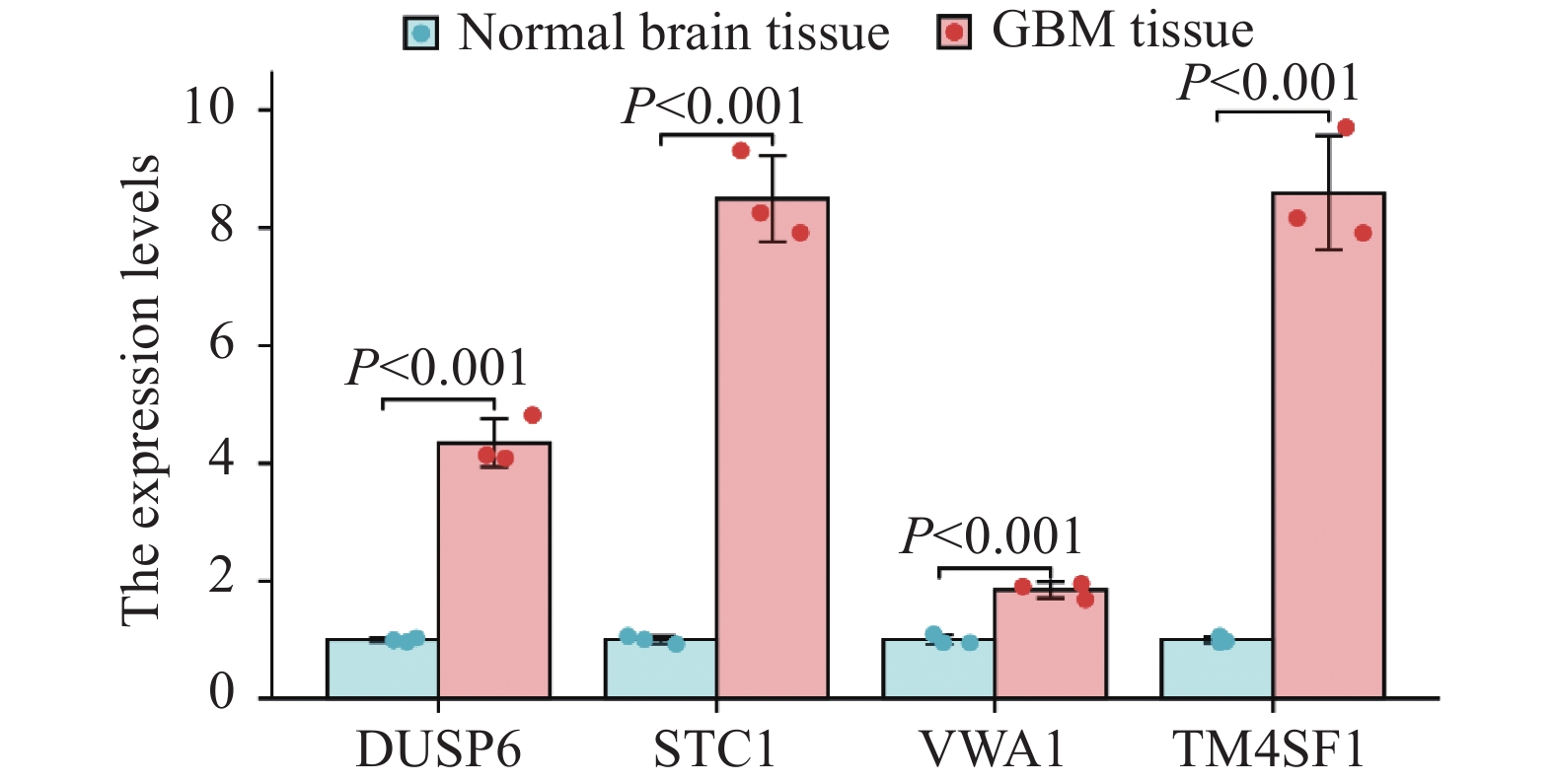
结果共筛选获得2 115个GBM预后相关基因,其中1 494个上调,621个下调。GBM单细胞测序分析降维后共获得7群细胞,分别为AC样肿瘤细胞、内皮细胞、单核/巨噬细胞、NB样肿瘤细胞、神经元、OC样肿瘤细胞以及OPC样肿瘤细胞。根据内皮细胞差异基因及相应筛选标准,经分析后共筛选出4个基因(DUSP6、STC1、VWA1和TM4SF1)用于风险评分的构建,并经qPCR实验验证目的基因在GBM组织及瘤周正常脑组织中的表达均显著上调。风险评分=0.171*DUSP6+0.144*STC1+0.041*VWA1−0.004*TM4SF1。
结论本研究构建的胶质母细胞瘤内皮细胞风险评分能够较好地预测患者预后。
Abstract:ObjectiveTo explore and verify the prognostic value of endothelial cells in glioblastoma.
MethodsThrough bioinformatics analysis of the TCGA and CGGA databases, we screened endothelial cell-related markers in GBM single-cell data according to a series of criteria. Moreover, univariate Cox regression analysis was performed to obtain and screen endothelial cell prognosis-related markers and construct endothelial cell-related prognostic risk score. qPCR experiments was used to verify the differences in the expression of prognostic markers in GBM tissues and peritumoral normal brain tissues. Kaplan-Meier method was used to construct the survival curve to identify the prognostic efficacy of the prognostic risk score.
ResultsA total of 2 115 prognostic genes of glioblastoma (GBM) were screened. Among them, 1 494 was upregulated and 621 was downregulated. Seven groups of cells were obtained after GBM single-cell sequencing analysis, including AC-like tumor cells, endothelial cells, monocytes/macrophages, NB-like tumor cells, neurons, OC-like tumor cells, and OPC-like tumor cells. According to the differential genes of endothelial cells and the corresponding screening criteria, four genes (DUSP6, STC1, VWA1, and TM4SF1) were screened for risk-score construction. The expression of the target gene in GBM tissues and normal brain tissues around the tumor was significantly up-regulated detected by qPCR. The risk score=0.171*DUSP6+0.144*STC1+0.041*VWA1−0.004*TM4SF1.
ConclusionThe glioblastoma endothelial cells’ risk score determined in this study can preferably predict the prognosis of patients.
-
Key words:
- Glioblastoma /
- Endothelial cell /
- Prognosis /
- Single-cell sequencing
-
Competing interests: The authors declare that they have no competing interests.利益冲突声明:所有作者均声明不存在利益冲突。作者贡献:马珩钞、刘羽阳:研究设计、数据收集、文章撰写许 俊:协助处理实验数据、补充研究设计缺陷陶丙岩:提供建议并协助实验操作张 军:指导研究设计、实施及论文撰写
-
表 1 qPCR实验所用引物
Table 1 Primers used in qPCR experiments
Gene name Gene ID Forward primer Reverse primer DUSP6 1 848 5′-GAACTGTGGTGTCTTGGTACATT-3′ 5′-GTTCATCGACAGATTGAGCTTCT-3′ STC1 6781 5′-CACGAGCTGACTTCAACAGGA-3′ 5′-GGATGTGCGTTTGATGTGGG-3′ VWA1 64856 5′-GCAGACTCGGGCTACTATGTG-3′ 5′-CACGTTGGACTCAGGCACTA-3′ TM4SF1 4071 5′-TGTGGCAAACGATGTGCGA-3′ 5′-TGACACAGTAGCCAGATCCTG-3′ 表 2 分析及数据可视化所用R包
Table 2 R packages for analysis and data visualization
Analysis item R package used LASSO regression
analysisglmnet (4.1.7) Single factor Cox
regression analysissurvival (3.3.1) and rms (6.3-0) Survival analysis survival (3.3.1), survminer and
ggplot2 (3.3.6)Venn diagram ggplot2 (3.3.6), VennDiagram (1.7.3) Forest plot ggplot2 (3.3.6) Co-expression heat map ggplot2 (3.3.6) -
[1] Le Rhun E, Preusser M, Roth P, et al. Molecular targeted therapy of glioblastoma[J]. Cancer Treat Rev, 2019, 80: 101896. doi: 10.1016/j.ctrv.2019.101896
[2] Grochans S, Cybulska AM, Simińska D, et al. Epidemiology of Glioblastoma Multiforme-Literature Review[J]. Cancers (Basel), 2022, 14(10): 2412. doi: 10.3390/cancers14102412
[3] Liu YY, Yao RQ, Long LY, et al. Worldwide productivity and research trend of publications concerning glioma-associated macrophage/microglia: A bibliometric study[J]. Front Neurol, 2022, 13: 1047162. doi: 10.3389/fneur.2022.1047162
[4] Ahir BK, Engelhard HH, Lakka SS. Tumor Development and Angiogenesis in Adult Brain Tumor: Glioblastoma[J]. Mol Neurobiol, 2020, 57(5): 2461-2478. doi: 10.1007/s12035-020-01892-8
[5] Hovis G, Chandra N, Kejriwal N, et al. Understanding the Role of Endothelial Cells in Glioblastoma: Mechanisms and Novel Treatments[J]. Int J Mol Sci, 2024, 25(11): 6118.
[6] Sadhwani N, Aggarwal A, Mishra A, et al. Temporal muscle thickness as an independent prognostic marker in glioblastoma patients-a systematic review and meta-analysis[J]. Neurosurg Rev, 2022, 45(6): 3619-3628. doi: 10.1007/s10143-022-01892-3
[7] Tan AC, Ashley DM, López GY, et al. Management of glioblastoma: State of the art and future directions[J]. CA Cancer J Clin, 2020, 70(4): 299-312. doi: 10.3322/caac.21613
[8] Liu Y, Shi Y, Wu M, et al. Hypoxia-induced polypoid giant cancer cells in glioma promote the transformation of tumor-associated macrophages to a tumor-supportive phenotype[J]. CNS Neurosci Ther, 2022, 28(9): 1326-1338.
[9] Sareen H, Ma Y, Becker TM, et al. Molecular Biomarkers in Glioblastoma: A Systematic Review and Meta-Analysis[J]. Int J Mol Sci, 2022, 23(16): 8835. doi: 10.3390/ijms23168835
[10] Butler M, Pongor L, Su YT, et al. MGMT Status as a Clinical Biomarker in Glioblastoma[J]. Trends Cancer, 2020, 6(5): 380-391. doi: 10.1016/j.trecan.2020.02.010
[11] Śledzińska P, Bebyn MG, Furtak J, et al. Prognostic and Predictive Biomarkers in Gliomas[J]. Int J Mol Sci, 2021, 22(19): 10373.
[12] Bălașa A, Șerban G, Chinezu R, et al. The Involvement of Exosomes in Glioblastoma Development, Diagnosis, Prognosis, and Treatment[J]. Brain Sci, 2020, 10(8): 553. doi: 10.3390/brainsci10080553
[13] Dai L, Li Z, Liang W, et al. SOCS proteins and their roles in the development of glioblastoma[J]. Oncol Lett, 2022, 23(1): 5.
[14] Qin J, Jiang C, Cai J, et al. Roles of Long Noncoding RNAs in Conferring Glioma Progression and Treatment[J]. Front Oncol, 2021, 11: 688027.
[15] Chaudhary R. Potential of long non-coding RNAs as a therapeutic target and molecular markers in glioblastoma pathogenesis[J]. Heliyon, 2021, 7(3): e06502. doi: 10.1016/j.heliyon.2021.e06502
[16] Jarmuzek P, Kozlowska K, Defort P, et al. Prognostic Values of Systemic Inflammatory Immunological Markers in Glioblastoma: A Systematic Review and Meta-Analysis[J]. Cancers (Basel), 2023, 15(13): 3339.
[17] Setlai BP, Hull R, Reis RM, et al. MicroRNA Interrelated Epithelial Mesenchymal Transition (EMT) in Glioblastoma[J]. Genes (Basel), 2022, 13(2): 244. doi: 10.3390/genes13020244
[18] 田志, 贾薇, 王钊, 等. 固有免疫分子CD58在人脑胶质瘤中表达及意义的初步研究[J]. 国际神经病学神经外科学杂志, 2022, 49(3): 1-7. [Tian Z, Jia W, Wang Z, et al. A preliminary study on the expression and significance of the innate immune molecu-lar marker CD58 in human gliomas[J]. Guo Ji Shen Jing Bing Xue Shen Jing Wai Ke Xue Za Zhi, 2022, 49(3): 1-7.] Tian Z, Jia W, Wang Z, et al. A preliminary study on the expression and significance of the innate immune molecu-lar marker CD58 in human gliomas[J]. Guo Ji Shen Jing Bing Xue Shen Jing Wai Ke Xue Za Zhi, 2022, 49(3): 1-7.
[19] Tamma R, Ingravallo G, Annese T, et al. Tumor Microenvironment and Microvascular Density in Human Glioblastoma[J]. Cells, 2022, 12(1): 11. doi: 10.3390/cells12010011
[20] van Tellingen O, Yetkin-Arik B, de Gooijer MC, et al. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment[J]. Drug Resist Updat, 2015, 19: 1-12. doi: 10.1016/j.drup.2015.02.002
[21] Arvanitis CD, Ferraro GB, Jain RK. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases[J]. Nat Rev Cancer, 2020, 20(1): 26-41.
[22] Downs-Canner SM, Meier J, Vincent BG, et al. B Cell Function in the Tumor Microenvironment[J]. Annu Rev Immunol, 2022, 40: 169-193. doi: 10.1146/annurev-immunol-101220-015603
[23] Dapash M, Hou D, Castro B, et al. The Interplay between Glioblastoma and Its Microenvironment[J]. Cells, 2021, 10(9): 2257. doi: 10.3390/cells10092257
[24] Huntington ND, Cursons J, Rautela J. The cancer-natural killer cell immunity cycle[J]. Nat Rev Cancer, 2020, 20(8): 437-454. doi: 10.1038/s41568-020-0272-z
-
期刊类型引用(2)
1. 朱信雪,许伟,谢雅萍,魏淑萍,解苗苗,苏茗羽. CAR-T细胞治疗肝细胞肝癌患者的双向式路径化护理与持续质量改进研究. 当代护士(上旬刊). 2025(03): 74-78 .  百度学术
百度学术
2. 张虹锐,肖明兴. 自主创新时代下药学专业教育与创新创业融合的探索与实践. 大学化学. 2024(12): 23-31 .  百度学术
百度学术
其他类型引用(0)




 下载:
下载: