Proteomics and Phosphoproteomics Analysis of Effect of Retinoic Acid-Induced Protein 16 Knockout on Human Colon Cancer Cells
-
摘要:目的
分析人结肠癌HCT116细胞敲除维甲酸诱导蛋白16(RAI16)后细胞内总蛋白及磷酸化蛋白质表达的差异,探究RAI16影响HCT116细胞蛋白质功能的可能机制及相关信号通路。
方法收集并提取HCT116 KO和WT细胞蛋白,SDS-PAGE检验蛋白提取效果。利用胰蛋白酶酶解蛋白后,标记肽段并进行质谱分析。对鉴定到的差异蛋白及差异磷酸化蛋白质利用GO数据库、KEGG数据库和STRING数据库进行生物信息学分析。
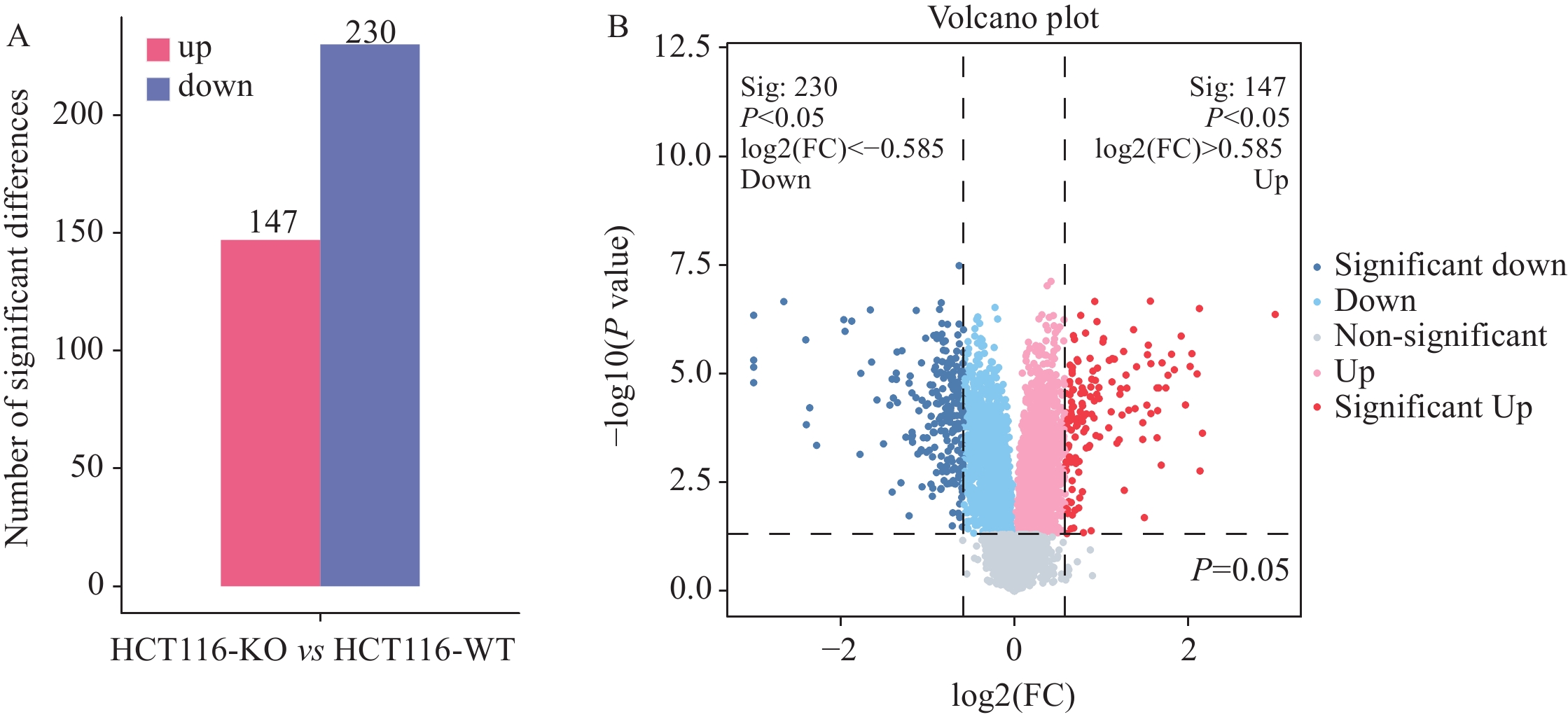
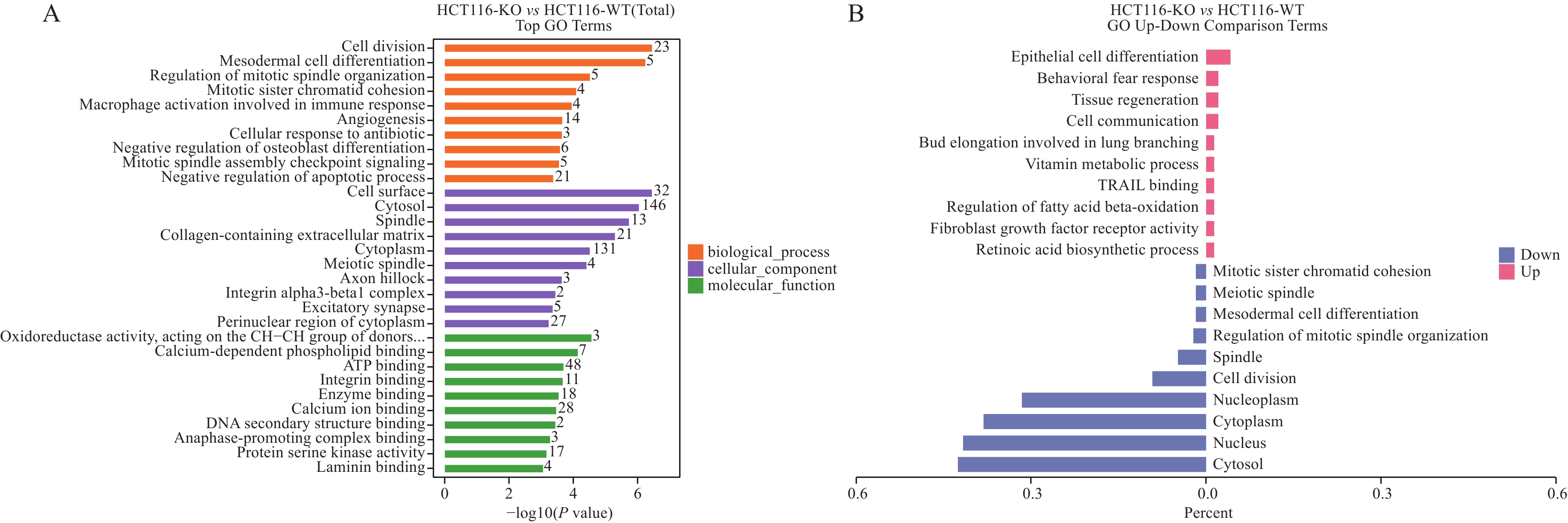
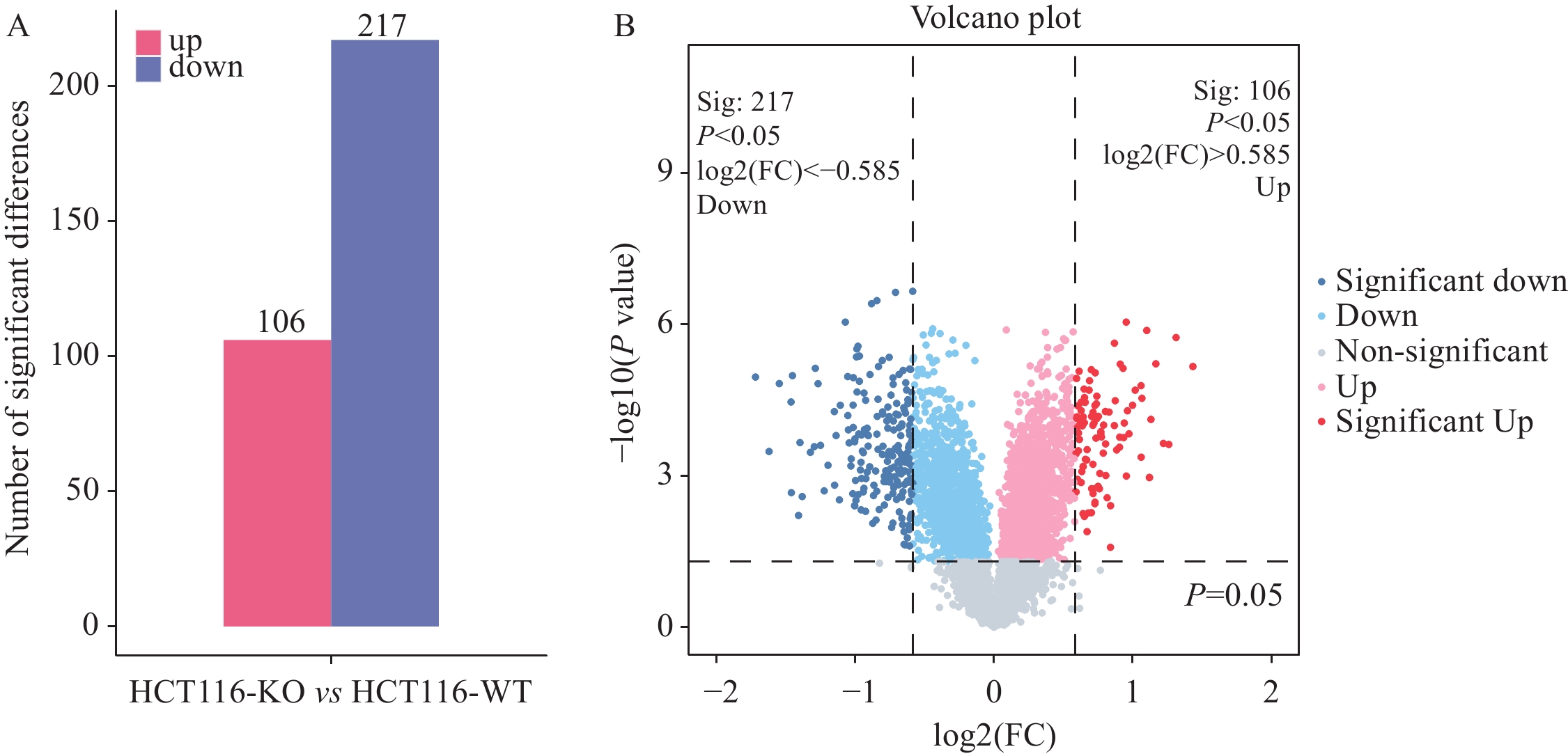
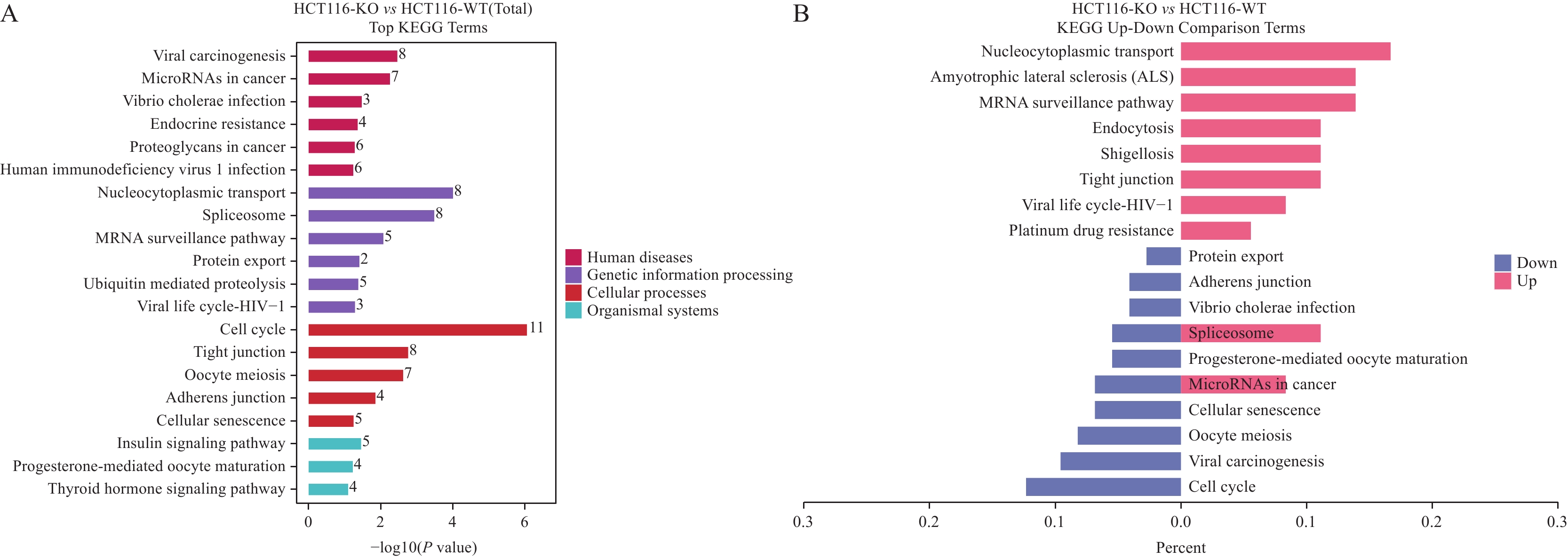
结果SDS-PAGE结果提示蛋白无明显降解,且实验组与对照组部分关键条带有明显差异;按Foldchange≥1.5或Foldchange≤1/1.5且P<0.05为条件进行差异蛋白的筛选,共筛选出147个上调差异蛋白和230个下调差异蛋白;并筛选到106个上调磷酸化位点和217个下调磷酸化位点。将去本底差异磷酸化位点功能GO富集分析,发现差异蛋白主要在核质、细胞核及细胞质组成,RNA、钙黏着蛋白及染色质结合,DNA修复、RNA剪接及DNA为模板转录的正调控等多个方面有显著富集趋势。KEGG富集结果显示,差异蛋白主要在核质转运、剪接体、细胞周期、细胞间紧密连接、病毒致癌作用和癌症中的微小RNA等通路具有显著富集趋势。蛋白互作网络主要以 DDX17、NCL、EEF2、CDK1、SSRP1和SMARCC1为核心蛋白质。统计发现,SKP1、ORC1和BAD等两组学差异变化均上调,且磷酸化差异变化比蛋白差异变化更显著,RBL1、RB1、CDK1、CDC6、MCM4、TFDP1、CHD4和SNW1等两组学差异变化均下调,且磷酸化差异变化比蛋白差异变化更显著。
结论RAI16可能通过SKP1、ORC1、RB1和CDK1等关键蛋白质在多方面生物功能和多条信号通路中发挥作用,影响细胞周期,进而影响癌症发生发展。
Abstract:ObjectiveTo analyze the differences in the expressions of the total and phosphorylated proteins in human colon cancer HCT116 cells after the knockout (KO) of retinoic acid-induced protein 16 (RAI16) and explore the possible mechanism and related signaling pathways affecting its protein function in HCT116 cells.
MethodsHCT116 KO and WT cell proteins were collected and extracted, and the protein extraction efficiency was detected via a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) experiment. After protein digestion, the peptides were labeled with TMT and analyzed via mass spectrometry. We used bioinformatics methods to analyze the identified differential proteins and differentially phosphorylated proteins by using GO, KEGG, and STRING databases.
ResultsThe results of SDS-PAGE showed no evident protein degradation. In addition, some key bands were significantly different between the experimental and control groups. A total of 147 up-regulated and 230 down-regulated differential proteins were screened in accordance with the conditions of Foldchange≥1.5 or Foldchange≤1/1.5 and P<0.05. Meanwhile, 106 up-regulated and 217 down-regulated phosphorylation sites were screened. GO enrichment analysis revealed that the differential proteins were mainly enriched in the composition of nucleoplasm, nucleus and cytoplasm, RNA binding, cadherin and chromatin, DNA repair, RNA splicing, and positive regulation of DNA as template transcription. The results of KEGG enrichment indicated that the differential proteins were mainly enriched in nucleocytoplasmic transport, spliceosomes, cell cycle, cell-cell tight junctions, viral carcinogenesis, microRNAs in cancer, etc. The protein interaction network mainly focused on DDX17, NCL, EEF2, CDK1, SSRP1, and SMARCC1. The statistical findings unveiled the up-regulated changes in the two omics of SKP1, ORC1, and BAD and the down-regulated changes in RBL1, RB1, CDK1, CDC6, MCM4, TFDP1, CHD4, and SNW1. Moreover, the phosphorylation differences were more significant than the protein differences.
ConclusionRAI16 plays the possible crucial role in multiple biological functions and signaling pathways through key proteins, such as SKP1, ORC1, RB1, and CDK1, which affect the cell cycle and thereby the occurrence and development of cancer.
-
Key words:
- Colon cancer /
- Retinoic acid-induced protein 16 /
- HCT116 cells /
- Phosphoproteomics
-
0 引言
胃癌(gastric cancer, GC)是严重威胁人类健康的恶性肿瘤之一。我国是胃癌高发国家,2020年胃癌新发病例数47.9万,死亡37.4万,约占全世界胃癌新发和死亡病例的44.0%和48.6%,防控形势十分严峻[1]。胃癌防治的最终目的是降低胃癌的发病率和死亡率,延长生命。由于目前对胃癌发生的复杂生物学过程和因素的理解尚不完全清楚,从根本上阻断致癌因素作用控制胃癌的发生在短时期难以突破。因此采用早期发现、早期诊断和早期治疗是降低胃癌患者死亡率、提高生存率和生存质量的重要途径。目前,胃镜是诊断胃癌的有效方法,但因其属侵入性检查,人群接受度较低,且费用较高,难以进行大规模普查。因此,急切需要开发一种可应用于胃癌早期筛查的简单可靠、无创或微创的大通量检查方法。多项研究表明,Dickkopf相关蛋白1(DKK1)在健康人群体内呈低表达,在肺癌、宫颈癌、肝癌、胰腺癌和胃癌等多种肿瘤患者中呈高表达,提示其可能作为一种潜在的肿瘤筛查标志物[2-6]。本研究探讨DKK1在胃癌患者和非胃癌者血清中的差异,并探索其与胃癌发生发展的关系,为评估其能否作为胃癌血清学筛查指标提供参考。
1 资料与方法
1.1 临床资料
选取2013年8月—2014年10月于中国医学科学院北京协和医学院肿瘤医院就诊、年龄范围40~69岁、经内镜检查及组织病理确诊为胃癌的患者170例,作为胃癌组。对照组来自同期本院体检科接受上消化道内镜检查且未诊断患胃癌者,且排除患有任何恶性肿瘤者,按照年龄(相差5岁以内)与胃癌病例进行随机1:1频数匹配。本研究经医院伦理委员会批准,所有受试者均签署知情同意书。所有研究对象的血液均为经过临床检测之后的剩余血标本,分离出1.5 ml血清,放入冻存管中,-80℃保存备用。采集血液时胃癌患者未经过放化疗或手术治疗。
1.2 检测方法
血清DKKl检测采用酶联免疫吸附实验(ELISA)。ELISA检测系统为美国R & D Systems公司生产的DKK100试剂盒,严格按照说明书进行操作。将100 μl的标准液或血清样品加入96孔酶标板中,室温孵育2 h,洗板。每孔加入200 μl DKK1结合液,室温孵育2 h,洗板。每孔加入200 μl的底物溶液,室温下避光孵育30 min,显色后加入50 μl的终止溶液并使用酶标仪检测450 nm处的光密度(optical density, OD)值。
1.3 统计学方法
采用SAS9.4进行数据整理与统计分析。血清DKK1浓度资料呈偏态分布,以中位数(四分位数)[M(P25, P75)]表示,两组间比较采用Wilcoxon秩和检验。分类资料以n(%)表示,采用Pearson χ2检验进行组间比较。血清DKK1浓度与胃癌患者的临床病理指标间的相关性比较采用Wilcoxon秩和检验或Kruskal-Wallis秩和检验。计算血清DKK1诊断胃癌的敏感度和特异性,绘制受试者工作特征ROC曲线,通过计算约登指数(Youden index)获取最佳诊断临界值。P < 0.05为差异有统计学意义。
2 结果
2.1 胃癌组与对照组的基本特征及血清DKK1浓度分布情况
胃癌组男性119例(70.00%),中位年龄59岁,对照组男性80例(47.06%),中位年龄58岁。两组在BMI、吸烟和饮酒的分布上差异无统计学意义(P > 0.05)。胃癌组患者血清DKK1的中位表达水平[266.4(182.1, 360.8)pg/ml]显著高于对照组[97.9(36.0, 145.1)pg/ml](P < 0.0001),见表 1。
表 1 胃癌组与对照组的基本特征及血清DKK1表达水平Table 1 Basic characteristics and expression of serum DKK1 in gastric cancer (GC) group and control group
2.2 血清DKK1在胃癌患者中的诊断意义
根据DKK1在胃癌组和对照组血清中的检测结果绘制ROC曲线,见图 1,曲线下面积为0.908(95%CI: 0.878~0.938)。根据约登指数最大确定诊断胃癌的最佳截断值为167.8 pg/ml。以DKK1浓度 > 167.8 pg/ml为DKK1阳性,诊断胃癌的敏感度为80.59%(95%CI: 76.64%~86.53%),特异性为84.71%(95%CI: 79.30%~90.12%),阳性预测值为84.05%(95%CI: 78.43%~89.67%),阴性预测值为81.36%(95%CI: 75.62%~87.09%)。
2.3 血清DKK1与胃癌患者临床病理指标的关系
血清DKK1与胃癌患者临床病理指标的关系见表 2。未发现血清DKK1浓度与胃癌患者的年龄、性别、民族、胃癌家族史、吸烟史、BMI、饮酒史、肿瘤大小、肝脏转移、血管浸润和远处转移之间存在相关性(均P > 0.05),但与肿瘤TNM分期、浸润深度、淋巴结转移和嗜神经侵袭明显相关(均P < 0.05)。
表 2 血清DKK1水平与胃癌患者临床病理特征的关系(M, P25-P75)Table 2 Relation between serum DKK1 level and clinicopathological characteristics of gastric cancer patients (M, P25-P75)
2.4 血清DKK1对胃癌患者临床病理特征的诊断价值
经ROC曲线分析,血清DKK1对胃癌浸润深度、淋巴结转移、嗜神经侵袭和TNM分期均具有一定的诊断意义(AUC=0.633、0.622、0.613、0.635)。根据约登指数,区分胃癌浸润深度(T3+T4 vs. T1+T2)、是否存在淋巴结转移、是否存在嗜神经侵袭、TNM分期(Ⅲ+Ⅳ vs.Ⅰ+Ⅱ)的最佳临界值分别为265.7、240.4、198.2、265.7 pg/ml,见表 3。
表 3 血清DKK1对胃癌临床病理特征的诊断价值Table 3 Diagnosis value of serum DKK1 on clinicopathological features of GC patients
3 讨论
21世纪以来,我国胃癌的发病率和死亡率均有所下降,生存率有所提高,但仍远低于日本与韩国等同样胃癌高发的国家,2010—2014年我国胃癌年龄标化5年生存率为35.9%,日本为60.3%,韩国最高,达到了68.9%[7]。而日本与韩国生存率提高的重要原因之一在于其所开展的全国性胃癌筛查项目。我国人口众多,目前还无法实现全民胃癌内镜普查,故通过胃癌的危险因素和一些标志物筛选高危人群进行内镜检查是符合我国国情的筛查策略[8]。目前,已知的胃癌血清标志物有胃蛋白酶原(PG)、胃泌素-17(G-17)、幽门螺杆菌抗体(Hp-IgG)、癌胚抗原(CEA)、CA199、CA724、微小RNA和环状RNA等,但尚没有一种被证实可作为胃癌筛查可靠的标志物[9]。2019年早期胃癌筛查专家共识中提出了一种新型胃癌筛查评分系统,通过性别、年龄与传统的血清学标志物PG、G-17和Hp-IgG五项指标浓缩高危人群行内镜精查,但其在真实世界人群中的筛查效果如何仍有待进一步研究评估[8]。故探索新的胃癌血清学标志物,对胃癌高危人群的筛选具有重要意义。
DKK1作为一种分泌蛋白,是有力的Wnt信号通路的拮抗剂[10]。Wnt信号通路为进化上非常保守的信号转导途径,在胚胎发育形成和维持成体组织的动态平衡中发挥重要作用,是调控细胞形态、生长、发育、增殖、迁移和分化的关键通路之一[11]。与正常干细胞一样,肿瘤干细胞也利用Wnt信号通道,这些通道的失调可能会导致肿瘤发生[12]。有研究发现,在正常胃腺和肠化生组织中,DKK1呈阴性或较低水平,而在胃癌细胞中通常呈中等或高度表达[6]。
本研究结果表明胃癌组血清DKK1的水平(266.4 pg/ml)显著高于对照组(97.9 pg/ml)(P < 0.0001)。多项研究也表明胃癌患者血清DKK1水平高于对照人群,提示血清DKK1具有作为胃癌筛查标志物的潜能[6, 13-16]。Lee等[6]对153例胃癌患者和173例健康对照者的血清DKK1进行检测,结果发现胃癌患者血清DKK1水平显著高于健康对照者(P < 0.0001)。唐丹等[13]比较了胃癌组、胃良性疾病组和健康对照组的血清DKK1水平,结果显示胃癌组 > 胃良性疾病组 > 健康对照组(P < 0.05),对胃癌具有较高的诊断价值(AUC=0.820)。Zhuang等[17]评估了90例胃癌患者、50例胃癌前疾病者与40例健康对照者的血清DKK1浓度,结果表明胃癌患者组血清DKK1浓度显著高于胃癌前疾病组和健康对照组(P < 0.01)。魏礼清等[14]对78例胃癌患者及38例健康对照者血清DKK1浓度进行检测,结果显示胃癌组平均水平(51.62±38.43 μg/ml)显著高于健康对照组(22.18±6.59 μg/ml),与本研究结果一致。而张小刚等[18]对34例胃癌患者和38例健康者血清DKK1检测的结果显示,胃癌患者血清中DKKl含量明显低于健康者(P < 0.01)。这与以上研究结果完全相反,可能是由于该研究样本量较小。
本研究还发现肿瘤分期晚、浸润深、存在神经侵袭与淋巴结转移者血清DKK-1水平更高,提示血清DKK1水平与胃癌恶性程度相关,与已发表的研究结果一致[17, 19-20]。Zhuang等[17]进一步研究发现血清DKK1水平与胃癌患者临床分期、血管浸润、浸润深度有关。卢起飞等[20]纳入132例胃癌患者、121例慢性胃炎患者及74例体检健康者的研究发现胃癌患者血清DKK-1水平与淋巴结转移、肿瘤分期有关(P < 0.05),不过该研究还发现血清DKK-1水平与肿瘤直径、肝脏转移有关,而未发现与肿瘤分化程度、浸润深度、血管转移情况相关(P > 0.05)。本研究并未发现血清DKK-1水平与肿瘤直径、肝脏转移、血管浸润有关(P > 0.05),考虑可能是发生肝脏转移和血管浸润的患者数较少或者受幽门螺杆菌感染、胃癌分型等混杂因素的影响,但仍需进一步研究验证。
本研究的局限性在于:(1)DKK1是一种非特异性的肿瘤标志物,并不适合单独用于胃癌筛查,而本研究仅分析了胃癌患者血清DKK1的表达水平,需结合PG、Hp-IgG等现有的胃癌血清标志物,进一步研究DKK1的加入是否可提高筛查的准确性;(2)本研究为一项单中心研究,未开展外部验证;(3)本研究胃癌患者与非胃癌者的性别构成存在差异,虽然按性别的分层分析显示男女血清DKK1浓度的曲线下面积比较接近,但对研究结果仍可能存在影响。今后将进一步完善研究方案,选择更具代表性的人群,以便更好地评估DKK1在胃癌筛查中的价值。
综上所述,血清DKK1作为胃癌标志物具有一定的价值,应开展进一步研究,探讨其在识别胃癌高危人群方面的作用。
Competing interests: The authors declare that they have no competing interests.利益冲突声明:所有作者均声明不存在利益冲突。作者贡献:陈奕孛:收集标本、实施实验、论文撰写苗 根:收集标本、标本前处理、数据整理王 文:设计实验、结果分析、论文修改丁翠玲、戚中田:设计实验、论文审校 -
表 1 HCT116细胞敲除RAI16后差异表达显著的关键蛋白质
Table 1 Differentially expressed key proteins after RAI16 knockout in HCT116 cells
Gene name Product Regulation STAT1 Signal transducer and activator of transcription 1-alpha/beta Up CYP24A1 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial Up ITGB1 Integrin beta-1 Down DAPK1 Death-associated protein kinase 1 Up FGFR2 Fibroblast growth factor receptor 2 Up SMAD3 Mothers against decapentaplegic homolog 3 Up CDK6 Cyclin-dependent kinase 6 Down NOTCH1 Neurogenic locus notch homolog protein 1 Down CDKN1A Cyclin-dependent kinase inhibitor 1 Up RAD51 DNA repair protein RAD51 homolog 1 Down 表 2 HCT116细胞敲除RAI16后差异表达显著的关键磷酸化蛋白质
Table 2 Differentially expressed key phosphorylated proteins after RAI16 knockout in HCT116 cells
Gene name Product Regulation CDK1 Cyclin-dependent kinase 1 Down SKP1 S-phase kinase-associated protein 1 Up ORC1 Origin recognition complex subunit 1 Up CDC20 Cell division cycle protein 20 homolog Down RB1 Retinoblastoma-associated protein Down PTTG1 Securin Down RBL1 Retinoblastoma-like protein 1 Down 表 3 HCT116细胞敲除RAI16后差异表达显著的去本底蛋白修饰位点
Table 3 Differentially expressed key modified sites in background-removal proteins after RAI16 knockout in HCT116 cells
Gene name class Product NEGG description SKP1 Phospho-more up S-phase kinase-associated protein 1 Modification-dependent protein catabolic process ORC1 Phospho-more up Origin recognition complex subunit 1 DNA replication RBL1 Phospho-more down Retinoblastoma-like protein 1 Regulation of cell cycle RB1 Phospho-more down Retinoblastoma-associated protein Regulation of cell cycle CDK1 Phospho-more down Cyclin-dependent kinase 1 Cyclin-dependent protein serine/threonine kinase activity CDC6 Phospho-more down Cell division control protein 6 homolog DNA replication initiation MCM4 Phospho-more down DNA replication licensing factor MCM4 DNA replication initiation TFDP1 Phospho-more down Transcription factor Dp-1 Cell cycle CHD4 Phospho-more down Chromodomain-helicase-DNA-binding protein 4 Peripheral T cell tolerance induction BAD Phospho-more up Bcl2-associated agonist of cell death Positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress SNW1 Phospho-more down SNW domain-containing protein 1 Generation of catalytic spliceosome for second transesterification step -
[1] Chang YT, Kowalczyk M, Fogerson PM, et al. Loss of Rai1 enhances hippocampal excitability and epileptogenesis in mouse models of Smith-Magenis syndrome[J]. Proc Natl Acad Sci U S A, 2022, 119(43): e2210122119. doi: 10.1073/pnas.2210122119
[2] Nishikawa S, Uemoto Y, Kim TS, et al. Low RAI2 expression is a marker of poor prognosis in breast cancer[J]. Breast Cancer Res Treat, 2021, 187(1): 81-93. doi: 10.1007/s10549-021-06176-w
[3] Jiao Y, Li S, Gong J, et al. Comprehensive analysis of the expression and prognosis for RAI2: A promising biomarker in breast cancer[J]. Front Oncol, 2023, 13: 1134149. doi: 10.3389/fonc.2023.1134149
[4] Lou X, Deng W, Shuai L, et al. RAI2 acts as a tumor suppressor with functional significance in gastric cancer[J]. Aging (Albany NY), 2023, 15(21): 11831-11844.
[5] Zhang W, Kong L, Zhu H, et al. Retinoic Acid-Induced 2 (RAI2) Is a Novel Antagonist of Wnt/β-Catenin Signaling Pathway and Potential Biomarker of Chemosensitivity in Colorectal Cancer[J]. Front Oncol, 2022, 12: 805290. doi: 10.3389/fonc.2022.805290
[6] Besler K, Węglarz A, Keller L, et al. Expression Patterns and Corepressor Function of Retinoic Acid-induced 2 in Prostate Cancer[J]. Clin Chem, 2022, 68(7): 973-983. doi: 10.1093/clinchem/hvac073
[7] Yang C, Mou Z, Zhang Z, et al. Circular RNA RBPMS inhibits bladder cancer progression via miR-330-3p/RAI2 regulation[J]. Mol Ther Nucleic Acids, 2021, 23: 872-886. doi: 10.1016/j.omtn.2021.01.009
[8] Huang M, Ding J, Wu X, et al. EZH2 affects malignant progression and DNA damage repair of lung adenocarcinoma cells by regulating RAI2 expression[J]. Mutat Res, 2022, 825: 111792. doi: 10.1016/j.mrfmmm.2022.111792
[9] Yu Y, Lu X, Yan Y, et al. The lncRNA KIF9-AS1 Accelerates Hepatocellular Carcinoma Growth by Recruiting DNMT1 to Promote RAI2 DNA Methylation[J]. J Oncol, 2022, 2022: 3888798.
[10] El Gammal AT, Melling N, Reeh M, et al. High levels of RAI3 expression is linked to shortened survival in esophageal cancer patients[J]. Exp Mol Pathol, 2019, 107: 51-56. doi: 10.1016/j.yexmp.2019.01.013
[11] Jin C, Zhu Y, Wu Y, et al. RAI3 knockdown enhances osteogenic differentiation of bone marrow mesenchymal stem cells via STAT3 signaling pathway[J]. Biochem Biophys Res Commun, 2020, 524(2): 516-522. doi: 10.1016/j.bbrc.2020.01.098
[12] Yan X, Zhang M, Li B, et al. RAI14 Regulated by circNFATC3/miR-23b-3p axis Facilitates Cell Growth and Invasion in Gastric Cancer[J]. Cell Transplant, 2021, 30: 9636897211007055.
[13] Wang J, Cai Y, Luo J, et al. RAI14 silencing suppresses progression of esophageal cancer via the STAT3 pathway[J]. Aging (Albany NY), 2020, 12(18): 18084-18098.
[14] Ding CL, Qian CL, Qi ZT, et al. Identification of retinoid acid induced 16 as a novel androgen receptor target in prostate cancer cells[J]. Mol Cell Endocrinol, 2020, 506: 110745. doi: 10.1016/j.mce.2020.110745
[15] Wang W, Ding CL, Wu MX, et al. RAI16 maintains intestinal homeostasis and inhibits NLRP3-dependent IL-18/CXCL16-induced colitis and the progression of colitis-associated colorectal cancer[J]. Clin Transl Med, 2022, 12(8): e993. doi: 10.1002/ctm2.993
[16] Xu YL, Ding CL, Qian CL, et al. Retinoid acid induced 16 deficiency aggravates colitis and colitis-associated tumorigenesis in mice[J]. Cell Death Dis, 2019, 10(12): 958. doi: 10.1038/s41419-019-2186-9
[17] Thompson LL, Rutherford KA, Lepage CC, et al. Aberrant SKP1 Expression: Diverse Mechanisms Impacting Genome and Chromosome Stability[J]. Front Cell Dev Biol, 2022, 10: 859582. doi: 10.3389/fcell.2022.859582
[18] Hussain M, Lu Y, Tariq M, et al. A small-molecule Skp1 inhibitor elicits cell death by p53-dependent mechanism[J]. iScience, 2022, 25(7): 104591. doi: 10.1016/j.isci.2022.104591
[19] Tian C, Lang T, Qiu J, et al. SKP1 promotes YAP-mediated colorectal cancer stemness via suppressing RASSF1[J]. Cancer Cell Int, 2020, 20(1): 579. doi: 10.1186/s12935-020-01683-0
[20] Wang B, Song J. Structural basis for the ORC1-Cyclin A association[J]. Protein Sci, 2019, 28(9): 1727-1733. doi: 10.1002/pro.3689
[21] Hossain M, Bhalla K, Stillman B. Multiple, short protein binding motifs in ORC1 and CDC6 control the initiation of DNA replication[J]. Mol Cell, 2021, 81(9): 1951-1969. e6.
[22] Mas AM, Goñi E, Ruiz de Los Mozos I, et al. ORC1 binds to cis-transcribed RNAs for efficient activation of replication origins[J]. Nat Commun, 2023, 14(1): 4447. doi: 10.1038/s41467-023-40105-3
[23] Okano-Uchida T, Kent LN, Ouseph MM, et al. Endoreduplication of the mouse genome in the absence of ORC1[J]. Genes Dev, 2018, 32(13-14): 978-990. doi: 10.1101/gad.311910.118
[24] Wu L, Chen H, Yang C. Origin recognition complex subunit 1(ORC1) is a potential biomarker and therapeutic target in cancer[J]. BMC Med Genomics, 2023, 16(1): 243. doi: 10.1186/s12920-023-01691-9
[25] Sheng Y, Wei J, Yu F, et al. A critical role of nuclear m6A reader YTHDC1 in leukemogenesis by regulating MCM complex-mediated DNA replication[J]. Blood, 2021, 138(26): 2838-2852. doi: 10.1182/blood.2021011707
[26] Byrne AB, García AG, Brahamian JM, et al. A murine model of dengue virus infection in suckling C57BL/6 and BALB/c mice[J]. Animal Model Exp Med, 2021, 4(1): 16-26. doi: 10.1002/ame2.12145
[27] Ventura E, Iannuzzi CA, Pentimalli F, et al. RBL1/p107 Expression Levels Are Modulated by Multiple Signaling Pathways[J]. Cancers (Basel), 2021: 13.
[28] Yao Y, Gu X, Xu X, et al. Novel insights into RB1 mutation[J]. Cancer Lett, 2022, 547: 215870. doi: 10.1016/j.canlet.2022.215870
[29] Wei R, Ren X, Kong H, et al. Rb1/Rbl1/Vhl loss induces mouse subretinal angiomatous proliferation and hemangioblastoma[J]. JCI Insight, 2019, 4(22): e127889. doi: 10.1172/jci.insight.127889
[30] Naert T, Dimitrakopoulou D, Tulkens D, et al. RBL1 (p107) functions as tumor suppressor in glioblastoma and small-cell pancreatic neuroendocrine carcinoma in Xenopus tropicalis[J]. Oncogene, 2020, 39(13): 2692-2706. doi: 10.1038/s41388-020-1173-z
[31] Nakajima R, Deguchi R, Komori H, et al. The TFDP1 gene coding for DP1, the heterodimeric partner of the transcription factor E2F, is a target of deregulated E2F[J]. Biochem Biophys Res Commun, 2023, 663: 154-162. doi: 10.1016/j.bbrc.2023.04.092
[32] Wang Q, Bode AM, Zhang T. Targeting CDK1 in cancer: mechanisms and implications[J]. NPJ Precis Oncol, 2023, 7(1): 58. doi: 10.1038/s41698-023-00407-7
[33] El Dika M, Dudka D, Kloc M, et al. CDC6 as a Key Inhibitory Regulator of CDK1 Activation Dynamics and the Timing of Mitotic Entry and Progression[J]. Biology (Basel), 2023, 12(6): 855.
[34] Lim N, Townsend PA. Cdc6 as a novel target in cancer: Oncogenic potential, senescence and subcellular localisation[J]. Int J Cancer, 2020, 147(6): 1528-1534. doi: 10.1002/ijc.32900
[35] Jia M, Zou X, Yin S, et al. CHD4 orchestrates the symphony of T and B lymphocytes development and a good mediator in preventing from autoimmune disease[J]. Immun Inflamm Dis, 2022, 10(7): e644. doi: 10.1002/iid3.644
[36] Chang CL, Huang CR, Chang SJ, et al. CHD4 as an important mediator in regulating the malignant behaviors of colorectal cancer[J]. Int J Biol Sci, 2021, 17(7): 1660-1670. doi: 10.7150/ijbs.56976
[37] Hagman JR, Arends T, Laborda C, et al. Chromodomain helicase DNA-binding 4 (CHD4) regulates early B cell identity and V(D)J recombination[J]. Immunol Rev, 2022, 305(1): 29-42. doi: 10.1111/imr.13054
[38] Boac BM, Abbasi F, Ismail-Khan R, et al. Expression of the BAD pathway is a marker of triple-negative status and poor outcome[J]. Sci Rep, 2019, 9(1): 17496. doi: 10.1038/s41598-019-53695-0
[39] Sherill-Rofe D, Raban O, Findlay S, et al. Multi-omics data integration analysis identifies the spliceosome as a key regulator of DNA double-strand break repair[J]. NAR Cancer, 2022, 4(2): zcac013. doi: 10.1093/narcan/zcac013
-
期刊类型引用(8)
1. 冯娟,王少怡,冯思佳,柏强善,陈安. 肝细胞癌患者的血清外泌体长链非编码RNA Airn表达及其预后预测价值. 国际消化病杂志. 2025(01): 49-55 .  百度学术
百度学术
2. 王善平,贺程程,李志军,孙嫣. 浅谈消化内科学术型硕士研究生科研能力的培养. 现代消化及介入诊疗. 2025(03): 393-397 .  百度学术
百度学术
3. 宋慧霞,宋盈花,谷申森. 无痛理念下认知行为干预对腹腔镜肝切除术患者疼痛评分及心理状态的影响. 中国临床研究. 2024(01): 141-146 .  百度学术
百度学术
4. 宋振宇,杨丽超,廖舟翔,张起,温莎,黄雪静,何敏. lnc85通过调节CDC42的表达促进肝癌细胞增殖. 广西医科大学学报. 2024(02): 186-192 .  百度学术
百度学术
5. 杨帆,段昌虎,吴林,赵李飞,向俊西,段建峰. 帕博利珠单抗联合乐伐替尼转化治疗后序贯根治性手术在初始不可切除肝癌中的应用效果. 中国药物应用与监测. 2024(04): 385-389 .  百度学术
百度学术
6. 菅辉玲,高丽霞,李桂圆,朱海鹏,李砥,贾晓玲,胡军. 联合检测血清APE1、miR183在原发性肝癌诊断中的价值. 肝脏. 2024(12): 1498-1502+1516 .  百度学术
百度学术
7. 张建花,章志翔,李蓄涵,曹惟嘉,耿佳琦,于成云,阎新宇. 基于银纳米颗粒免疫比色法检测甲胎蛋白. 化学试剂. 2023(10): 40-45 .  百度学术
百度学术
8. 成晓梅,田红,李想,杨洁. HBV相关肝细胞癌患者血清miR-504、miR-200a、IL-24水平及其与肝癌的相关性. 肝脏. 2023(10): 1186-1190 .  百度学术
百度学术
其他类型引用(1)



 下载:
下载: