Fanconi Anemia: Exploration of DNA Repair Pathways from Genetic Diseases to Cancer and Prospects for Treatment
-
摘要:
范可尼贫血(FA)是一种遗传性疾病,其特征包括骨髓衰竭、发育异常和易患癌症。这种疾病是由基因突变引起的,导致修复DNA链间交联(ICLs)异常。DNA损伤反应失调会导致基因组不稳定,增加突变率和致癌风险。FA通路是DNA损伤应答的重要组成部分,在DNA链间交联修复和基因组稳定性方面发挥着关键作用。任何一个编码FA蛋白的基因胚系突变都会导致FA。随着体细胞癌中FA基因表达异常的普遍发生和不断开展的FA通路激活与化疗耐药相关性的研究,FA通路与癌症之间的联系得到了进一步确认,并且基于FA通路基因缺陷的靶向治疗也在逐步开发和应用。本文综述了FA蛋白在ICLs修复、FA信号网络调节以及其在癌症发病和预后中的重要作用,并探讨了靶向FA途径的小分子抑制剂的潜在应用。
Abstract:Fanconi anemia (FA) is an inheritable disorder that presents with bone marrow failure, developmental anomalies, and an increased susceptibility to cancer. The etiology of this condition stems from a genetic mutation that disrupts the proper repair of interstrand DNA cross-links (ICLs). The resultant dysregulation of the DNA damage response mechanism can induce genomic instability, thereby elevating the mutation rates and the likelihood of developing cancer. The FA pathway assumes a pivotal role in safeguarding genome stability through its involvement in the repair of DNA cross-links and the maintenance of overall genomic integrity. A mutation in the germ line of any of the genes responsible for encoding the FA protein results in the development of FA. The prevalence of aberrant FA gene expression in somatic cancer, coupled with the identification of a connection between FA pathway activation and resistance to chemotherapy, has solidified the correlation between the FA pathway and cancer. Consequently, targeted therapies that exploit FA pathway gene abnormalities are being progressively developed and implemented. This review critically examines the involvement of the FA protein in the repair of ICLs, the regulation of the FA signaling network, and its implications in cancer pathogenesis and prognosis. Additionally, it explores the potential utility of small-molecule inhibitors that target the FA pathway.
-
Key words:
- Fanconi anemia pathway /
- DNA damage repair /
- Cancer susceptibility /
- Prognosis /
- Targeted therapy
-
0 引言
范可尼贫血(fanconi anemia, FA)是一种罕见的常染色体隐性遗传病,涉及多种类型的血细胞减少、先天性畸形和肿瘤性疾病[1]。目前对这一临床和生物学上复杂的癌症易感综合征的认识取得了显著进展。在主要通过细胞周期S期激活的FA通路中,DNA损伤修复受到大量蛋白之间相互作用的调节,包括22种FA相关蛋白。编码FA蛋白的基因中任何一个基因的遗传变异都会导致不同亚型的FA。当外源DNA损伤如顺铂等交联剂损伤导致复制受阻时,复制叉失活,从而激活FA通路(也称为FA-BRCA通路),通过磷酸化、泛素化等多种修饰募集FA相关蛋白。其中,FA核心复合物对FANCD2-FANCI异源二聚体的单泛素化促进DNA修复效应物募集到染色质损伤,以解决DNA损伤和有丝分裂[2]。此外,由于FA通路与癌症易感性有关,BRCA1和BRCA2等一些FA基因的单个等位基因胚系失活会导致常见的家族性乳腺癌易感综合征[3]。因此,更好地了解该通路的机制和作用将有助于开发更具针对性的癌症治疗药物。
1 链间交联与范可尼贫血通路的作用机制
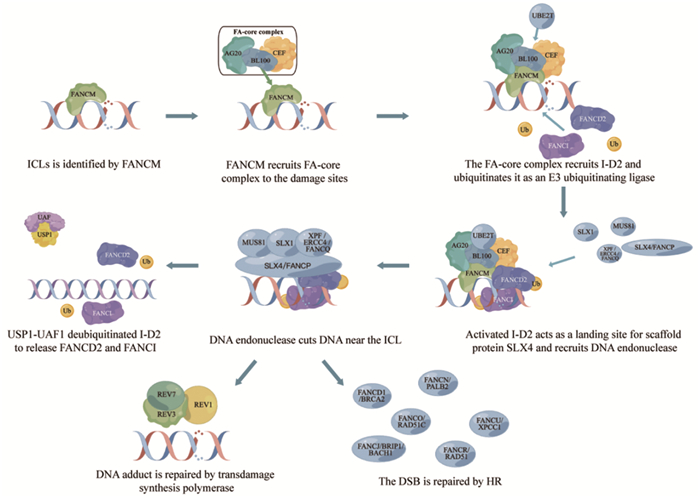
范可尼贫血是一种罕见的常染色体隐性遗传病,涉及多种类型的血细胞减少、先天性畸形和肿瘤性疾病[3]。FA患者的细胞表现出自发性基因组不稳定性,并且对DNA链间交联剂(如MMC和内源性醛类)敏感,也可能诱导DNA-蛋白质交联[4]。DNA链间交联(interstrand crosslinks, ICL)是由各种内源性代谢物、环境暴露和癌症化疗药物引起的一种病理变化,其共同特征是在两个核苷酸上形成共价连接,从而阻碍DNA的分离,使包括DNA复制和转录在内的基本细胞过程受到阻碍。ICL主要在DNA复制分叉停滞时的S阶段被检测到。范可尼贫血途径、DNA修复和染色质结构蛋白的大量翻译后修饰促进了ICL的损伤信号转导和修复。未修复的ICL会导致DNA断裂和染色体重排,这解释了范可尼贫血的骨髓衰竭特征以及为什么交联剂在癌症治疗中有效。目前已知22种FA基因(FANCA-W)的突变将会导致该疾病的发生[5-6]。这22个FA基因产物可分为四个功能组:第一组是FANCM复合体,即损伤传感模块;第二组是E3泛素连接酶,又称为FA核心复合物;第三组是FANCI-FANCD2(I-D2)异二聚体复合物,是FA通路和E3连接酶底物的关键效应物;第四组是下游FA蛋白,包括其他DNA修复/损伤耐受因子[7]。FA通路的一个关键功能是通过I-D2异二聚体的单泛素化和染色质靶向来协调DNA损伤反应(DNA damage response, DDR)。下文将对FA通路对ICLs的修复进行概述,见图 1。
1.1 损伤信号的识别与FA通路的激活
损伤信号转导始于将FANCM、FAAP24和MHF募集到复制叉中,与未缠绕的DNA结合[8]。FANCM对复制叉的重塑导致ssDNA结合蛋白,即复制蛋白A(replication protein A, RPA)的募集。RPA定位到DNA是ATR激活所必需的[9]。FANCM一旦与染色质结合,就可以作为FA核心复合物的着陆平台。FA核心复合物蛋白被招募到DNA损伤位点,与多种蛋白质相互作用,并作为范可尼贫血蛋白FANCD2和FANCI的泛素连接酶(E3泛素化连接酶)发挥作用[9]。
1.2 I-D2在FA通路中的重要作用
I-D2的激活至少涉及两种翻译后修饰。首先,在FA核心复合物募集到ICLs时,I-D2的磷酸化被激活[10]。FANCI和FANCD2随后以相互依赖的方式单泛素化。该单泛素化步骤由FA核心复合物通过其泛素连接酶亚基FANCL和相应的E2泛素结合酶UBE2T(又称FANCT)催化完成[11-13]。泛素化的FANCD2定位于ICL区,在ICL区充当SLX4和FA相关核酸酶1(Fanconi anemia-associated nuclease 1, FAN1)募集因子的着陆点[14-15]。同时,FANCD2协调DNA内切酶MUS81、SLX1和XPF/ERCC4/FANCQ的作用[16-17]。这些内切酶切割ICL附近的DNA链,产生DNA加合物和ICL的双链断裂[1]。DNA加合物由跨损伤合成聚合酶如REV1或DNA聚合酶ζ(即REV3-REV7)绕过,而ICL来源的双链断裂则通过HR修复[18]。在这个高度保守的途径中,DNA切口产生DSB,进一步DNA外切酶如CtBP相互作用蛋白(CtBP-interacting protein, CtIP)、MRN(MRE11-RAD50-NBS1)和核酸外切酶1(exonuclease 1, EXO1)切除双链断裂,从而产生3'单链DNA悬垂[19-20]。DNA悬垂最初被RPA包被,一旦RAD51/FANCR重组酶识别RPA覆盖的ssDNA,在BRCA2/FANCD1、FANCN/PALB2、RAD51C/FANCO、BRIP1/BACH1/FANCJ和XRCC1/FANCU蛋白的协助下,RAD51/FANCR重组酶促进RPA从3'悬垂中移除并刺激重组丝的形成。重组丝对最终的修复过程进行同源搜索并催化同源配对和DNA链侵袭和交换[21-22]。值得注意的是,ICLs的识别不仅导致FA通路修复过程,还导致ATR-CHK1细胞周期检查点的激活,从而减缓DNA复制并允许修复发生。
1.3 修复后FA通路的关闭
完成DNA损伤修复后需要关闭FA通路,以防止细胞周期阻滞延长和细胞死亡。进一步翻译和修饰FA蛋白可以促进终止FA通路反应。在DNA修复完成后,FANCD2-FANCI单泛素化被泛素羧基末端水解酶1(ubiquitin carboxyl-terminal hydrolase 1, USP1)- USP1相关因子1(USP1-associated factor 1, UAF1;又称WDR48)去泛素化酶复合物逆转[13, 23]。剩余的加合物通过核苷酸切除修复(nucleotide excision repair, NER)去除,以完成ICL修复[24]。此外,通过SUMO靶向泛素E2连接酶RNF3和ATPase p97及时从染色质中提取FANCD2,在调节DNA修复位点活性和FANCD2水平的平衡中起着重要作用。修复后关闭FA通路对于限制核酸酶活性以及促使ID复合物在DNA修复的多个步骤中发挥作用是必要的[25]。
2 范可尼贫血通路与癌症的相关性
2.1 FA患者的患癌风险增加
FA患者往往会出现造血功能衰竭,这与进行性全血细胞减少症有关。这导致缺乏功能性的骨髓干细胞,而骨髓干细胞是细胞周转过程中补充血细胞数量的必要条件。严重的FA患者会发展成急性骨髓性白血病,但随着造血干细胞移植的不断发展,FA患者的生存状况已明显改善,生育能力的降低和肿瘤发病率的增加已经成为年轻FA患者的严重问题[26]。造血干细胞移植增加了实体瘤的形成倾向,这与造血干细胞移植前的化疗方案(环磷酰胺和放疗)有关。环磷酰胺和放疗可促使ICL的发生,而由于FA患者FA修复通路的缺乏使癌症发病率增长迅速[1]。
基因组不稳定是癌细胞的一个重要特征,该特征可能源于DNA修复机制的失败,该机制基本上是作为一个肿瘤抑制网络来维护基因组的完整性和防止恶性肿瘤。FA与癌症易感性之间的联系已被充分证实,FA患者群体往往会表现出各种各样的癌症[27]。有研究表明近25%的FA患者会发生恶性肿瘤[27],最常见的是血液病,如骨髓增生异常综合征和急性髓细胞白血病[28]。在实体瘤中,鳞状细胞癌对FA患者构成了最高的风险,主要发生在头、颈和食管[1],可能是由于这些组织的复制潜力增加,促进了癌症的生长。除此之外,FA相关的癌症通常发生在乳腺、肝脏、胰腺、外阴、肛门和卵巢组织[1, 29]。
2.2 FA相关基因突变与癌症的相关性
FA相关基因尤其是FA核心复合物任何成分的基因突变,都会导致FANCI-FANCD2单泛素化缺陷和DNA损伤敏感性、出生缺陷、早发性骨髓衰竭和癌症倾向。最近FA蛋白的突变在FA患者群体之外的家族性和散发性癌症中也有报道。
Del Valle等通过下一代定制测序组合分析了总共1 021例遗传性癌症患者和194例对照,研究发现最常突变的基因是FANCA、FANCL和FANCM[30],其中与Figlioli研究的南欧队列一致的是,FANCM中c.5791C > T突变与雌激素受体ER阴性乳腺癌风险相关[31]。Gulnur等通过对来自哈萨克斯坦的125例(年龄17~50岁)早发性CRC患者的外周血DNA基因组进行二代测序,识别出FANCA、FANCD2和FANCI突变可能与结直肠癌有关[32]。同时,在对包含573例肠癌患者的中国队列测序中,发现FANCD2、FANCA、FANCG、FANCL和SLX4的突变是癌症的易感因素[33]。除此之外,FANCA突变携带者与乳腺癌和卵巢癌的患病风险有显著关联[30],FANCD1突变与卵巢癌、乳腺癌、前列腺癌、胃癌和胰腺癌有关[34-36]。FANCL突变与乳腺癌和白血病有关[30, 37]。FANCD2突变与乳腺癌、肺腺癌等多种癌症相关[38-39]。FANCC和FANCG与胰腺癌、卵巢癌、乳腺癌有关[40-41]。
2.3 FA相关基因的过表达与癌症的相关性
如上所述,FA基因的获得性突变发生在非FA的个体散发性癌症中,并且FA基因突变相关的易癌表型支持FA蛋白的肿瘤抑制功能。除了基因突变外,FA基因的拷贝数变化也常见于多种癌症中。Liu等分别使用TCGA和GTEx数据库中的肿瘤和组织对单个FA基因的差异表达分析也同样表明,大多数FA基因在各类癌症中过表达[42]。
以FANCD2为例,研究表明,在肺腺癌中FANCD2的高表达水平与较高的TNM分期、高肿瘤负荷、低化疗敏感性、低免疫评分、低生存率有关[43]。Zheng等发现FANCD2在子宫内膜癌中高表达,且较高的FANCD2表达与更深的肌层浸润和更多的错配修复状态相关。敲低癌细胞系中FANCD2表达抑制了恶性增殖和迁移能力,并赋予EC细胞对链间交联剂的敏感性[44]。在散发性乳腺癌中FANCD2的过表达与激素受体阴性、HER2扩增、p53表达升高、高级别和高增殖相关,且FANCD2/Ki-67免疫荧光双染的结果表明,FANCD2与Ki-67在浸润性乳腺癌细胞中共表达,并且二者呈显著正相关[38]。另外一项研究评估了50例结直肠癌活检中的FANCT/UBE2T蛋白水平与手术前未接受放疗或化疗患者的配对非癌性黏膜的比较,结果显示,结直肠癌标本中FANCT蛋白水平高于配对的非癌组织。同样,与UBE2T蛋白表达低的标本相比,具有高FANCT蛋白表达的肿瘤组织样本与较晚期的TNM分期和更差的总生存期相关[45]。由此可见,多项研究表明FA相关基因的过表达与较强的癌细胞迁移、增殖能力、较差的临床分期、较差的预后相关[29, 42, 46]。
综上所述,一方面,缺乏FA通路和损伤修复通路的重定向造成基因组的不稳定,导致突变、缺失和易位的积累,构成肿瘤克隆选择的起始点。另一方面,FA基因表达的升高和DNA损伤修复能力的提高有利于减轻伴随癌细胞快速增殖带来的过量DNA损伤和染色体异常[42]。FA通路的双相需求模型在癌症发生和发展的整个过程中发挥着重要作用。
3 靶向范可尼贫血通路在癌症治疗中的作用
3.1 FA途径与耐药性的关系
一系列诱导DNA损伤的化疗药物,如顺铂,是卵巢癌、睾丸癌和其他缺乏有效靶向药物治疗疾病的标准临床疗法[47]。耐药性是限制其应用的主要因素之一[42]。FA通路是修复顺铂导致的联检交联所需的DNA损伤反应通路,该通路的再激活可能是癌症治疗中获得性顺铂耐药的原因之一。研究表明,具有FANCF甲基化的卵巢癌细胞系对顺铂敏感,而这些细胞中的FANCF脱甲基化可恢复其耐药性[48],在携带FANCF基因异常去甲基化的细胞系亚群中也发现了类似的耐药性[48]。近期研究发现,在顺铂耐药的非小细胞肺癌中观察到FA相关基因FANCL和RAD18的上调,通过下调这些基因同样恢复了对顺铂的敏感性。类似的方法显示,siRNA介导的FA相关基因FANCL和RAD18以及HR相关基因BRCA1和BRCA2的沉默,通过抑制FA通路显著增强了A549/DR细胞对顺铂的敏感性[49]。同时,增强的FA通路激活也被证明与多发性骨髓瘤和胰腺癌中对马法兰的耐药性有关[42]。这些实验数据均提示了FA通路在化疗耐药中的重要作用以及开发FA靶向药物的必要性。
3.2 靶向FA通路的特异性抑制剂
近年来有关DNA损伤化疗药物的耐药性研究在广泛开展,其中针对FA通路的靶向药物可作为增敏剂克服肿瘤细胞的耐药性,从而增强化疗药物的抗肿瘤活性。SERPINB3是一种丝氨酸蛋白酶抑制剂,在多种鳞状细胞癌中高表达,并表现出显著的抗凋亡特性。Huang等基于HPV阳性头颈鳞癌顺铂敏感反应的RNA测序鉴定了SERPINB3,发现SERPINB3可以被HPV癌蛋白E6下调,并通过抑制DNA损伤修复来促进HPV阳性HNSCC中的顺铂敏感性,并基于此构建了内体PH响应纳米颗粒系统靶向敲低SERPINB3,通过抑制USP1表达抑制FA途径中的FANCD2-FANCI去泛素化,打断顺铂诱导的ICL修复通路,最终引发细胞凋亡[50]。Yuan等通过JC-1染色、碘化丙啶染色和蛋白质印迹实验证实,双硫仑可以抑制FA修复途径使铂类化疗药物在内的DNA损伤剂产生致敏作用,从而增强肿瘤细胞的凋亡[51]。有研究报道复制体蛋白And-1可以通过感知ICL并将FANCM/FAAP24复合物募集到停滞的复制叉中,其与FANCM复合物协调进行ICL修复。Zhang等研究发现抑制And-1的活化可以抑制卵巢癌的顺铂耐药性[52]。
除此之外,目前已报道多种药物通过抑制FANCD2增强癌细胞对顺铂的敏感性。Singh等研究表明,planispine A通过抑制FANCD2单泛素化和FANCD2复合体的形成,因此与顺铂联合作用时增加癌细胞的凋亡[53]。小梭菌乙醇提取物联合顺铂用于非小细胞肺癌、exportin 1 inhibitors(XPO1i)联合melphalan用于多发性骨髓瘤等,也均是通过类似的机制增强癌细胞对化疗药物的敏感性,成为联合化疗的新型抗癌药物[54-55]。
4 结语与展望
范可尼贫血是一种隐性遗传性疾病,除了骨髓衰竭和先天性异常的病理表现外,FA还与染色体异常和基因组不稳定有关,因此代表了癌症易感性的遗传易感特性。随着癌症中FA基因改变的普遍发生和FA通路激活相关化疗耐药性的观察,FA通路与癌症的相关性进一步确立。因此,明确FA通路修复ICLs并保护基因组完整性的详细机制具有重要意义,通过了解与FA相关的致癌机制,有望促进有效预防、治疗和早期检测范可尼贫血相关癌症的发展研究,并进一步开发针对FA和家族性或散发性癌症的个体化靶向治疗方案。
Competing interests: The authors declare that they have no competing interests.利益冲突声明:所有作者均声明不存在利益冲突。作者贡献:史晋宇:论文构思、撰写与修改邢琳:论文修改刘世佳、吕文豪、张冰琰、徐立君:绘图、校对张亚芬:论文整体思路设计 -
[1] Peake JD, Noguchi E. Fanconi anemia: current insights regarding epidemiology, cancer, and DNA repair[J]. Hum Genet, 2022, 141(12): 1811-1836. doi: 10.1007/s00439-022-02462-9
[2] Lemonidis K, Rennie ML, Arkinson C, et al. Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination[J]. EMBO J, 2023, 42(3): e111898. doi: 10.15252/embj.2022111898
[3] Narod SA. Which Genes for Hereditary Breast Cancer?[J]. N Engl J Med, 2021, 384(5): 471-473. doi: 10.1056/NEJMe2035083
[4] Hodskinson M R, Bolner A, Sato K, et al. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms[J]. Nature, 2020, 579(7800): 603-608. doi: 10.1038/s41586-020-2059-5
[5] Chen HZ, Li N, Wang J. Research progress of Fanconi anemia and DNA interstrand crosslink repair[J]. Zhonghua Xue Ye Xue Za Zhi, 2022, 43(2): 173-176.
[6] Webster ALH, Sanders MA, Patel K, et al. Genomic signature of Fanconi anaemia DNA repair pathway deficiency in cancer[J]. Nature, 2022, 612(7940): 495-502. doi: 10.1038/s41586-022-05253-4
[7] Ishiai M. Regulation of the Fanconi Anemia DNA Repair Pathway by Phosphorylation and Monoubiquitination[J]. Genes (Basel), 2021, 12(11): 1763. doi: 10.3390/genes12111763
[8] Nikfarjam S, Singh KK. DNA damage response signaling: A common link between cancer and cardiovascular diseases[J]. Cancer Med, 2023, 12(4): 4380-4404. doi: 10.1002/cam4.5274
[9] Ma M, Rodriguez A, Sugimoto K. Activation of ATR-related protein kinase upon DNA damage recognition[J]. Curr Genet, 2020, 66(2): 327-333. doi: 10.1007/s00294-019-01039-w
[10] Sijacki T, Alcon P, Chen ZA, et al. The DNA-damage kinase ATR activates the FANCD2-FANCI clamp by priming it for ubiquitination[J]. Nat Struct Mol Biol, 2022, 29(9): 881-890. doi: 10.1038/s41594-022-00820-9
[11] Yu Y, Xu W, Wen C, et al. UBE2T resolves transcription-replication conflicts and protects common fragile sites in primordial germ cells[J]. Cell Mol Life Sci, 2023, 80(4): 92. doi: 10.1007/s00018-023-04733-8
[12] Wang S, Wang R, Peralta C, et al. Structure of the FA core ubiquitin ligase closing the ID clamp on DNA[J]. Nat Struct Mol Biol, 2021, 28(3): 300-309. doi: 10.1038/s41594-021-00568-8
[13] Lemonidis K, Arkinson C, Rennie ML, et al. Mechanism, specificity, and function of FANCD2-FANCI ubiquitination and deubiquitination[J]. FEBS J, 2022, 289(16): 4811-4829. doi: 10.1111/febs.16077
[14] Aprosoff CM, Dyakov BJA, Cheung VHW, et al. Comprehensive Interactome Mapping of the DNA Repair Scaffold SLX4 Using Proximity Labeling and Affinity Purification[J]. J Proteome Res, 2023, 22(6): 1660-1681. doi: 10.1021/acs.jproteome.2c00706
[15] Elango R, Panday A, Lach FP, et al. The structure-specific endonuclease complex SLX4-XPF regulates Tus-Ter-induced homologous recombination[J]. Nat Struct Mol Biol, 2022, 29(8): 801-812. doi: 10.1038/s41594-022-00812-9
[16] Sabatella M, Thijssen KL, Davo-Martinez C, et al. Tissue-Specific DNA Repair Activity of ERCC-1/XPF-1[J]. Cell Rep, 2021, 34(2): 108608. doi: 10.1016/j.celrep.2020.108608
[17] Payliss BJ, Tse YWE, Reichheld SE, et al. Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease[J]. Cell Rep, 2022, 41(4): 111537. doi: 10.1016/j.celrep.2022.111537
[18] Rizzo AA, Vassel FM, Chatterjee N, et al. Rev7 dimerization is important for assembly and function of the Rev1/Polzeta translesion synthesis complex[J]. Proc Natl Acad Sci U S A, 2018, 115(35): E8191-E8200.
[19] Mozaffari NL, Pagliarulo F, Sartori AA. Human CtIP: A 'double agent' in DNA repair and tumorigenesis[J]. Semin Cell Dev Biol, 2021, 113: 47-56. doi: 10.1016/j.semcdb.2020.09.001
[20] Xu Y, Xu D. Repair pathway choice for double-strand breaks[J]. Essays Biochem, 2020, 64(5): 765-777. doi: 10.1042/EBC20200007
[21] Mekonnen N, Yang H, Shin YK. Homologous Recombination Deficiency in Ovarian, Breast, Colorectal, Pancreatic, Non-Small Cell Lung and Prostate Cancers, and the Mechanisms of Resistance to PARP Inhibitors[J]. Front Oncol, 2022, 12: 880643. doi: 10.3389/fonc.2022.880643
[22] Cong K, Peng M, Kousholt AN, et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency[J]. Mol Cell, 2021, 81(15): 3128-3144,e7. doi: 10.1016/j.molcel.2021.06.011
[23] Rennie ML, Arkinson C, Chaugule VK, et al. Structural basis of FANCD2 deubiquitination by USP1-UAF1[J]. Nat Struct Mol Biol, 2021, 28(4): 356-364. doi: 10.1038/s41594-021-00576-8
[24] Zhang X, Yin M, Hu J. Nucleotide excision repair: a versatile and smart toolkit[J]. Acta Biochim Biophys Sin (Shanghai), 2022, 54(6): 807-819. doi: 10.3724/abbs.2022054
[25] Martin-Rufo R, de la Vega-Barranco G, Lecona E. Ubiquitin and SUMO as timers during DNA replication[J]. Semin Cell Dev Biol, 2022, 132: 62-73. doi: 10.1016/j.semcdb.2022.02.013
[26] Vanni VS, Campo G, Cioffi R, et al. The neglected members of the family: non-BRCA mutations in the Fanconi anemia/BRCA pathway and reproduction[J]. Hum Reprod Update, 2022, 28(2): 296-311. doi: 10.1093/humupd/dmab045
[27] Altintas B, Giri N, McReynolds LJ, et al. Genotype-phenotype and outcome associations in patients with Fanconi anemia: the National Cancer Institute cohort[J]. Haematologica, 2023, 108(1): 69-82.
[28] Arai H, Minami Y, Chi S, et al. Molecular-Targeted Therapy for Tumor-Agnostic Mutations in Acute Myeloid Leukemia[J]. Biomedicines, 2022, 10(12): 3008. doi: 10.3390/biomedicines10123008
[29] Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship[J]. Nat Rev Cancer, 2018, 18(3): 168-185. doi: 10.1038/nrc.2017.116
[30] Del Valle J, Rofes P, Moreno-Cabrera JM, et al. Exploring the Role of Mutations in Fanconi Anemia Genes in Hereditary Cancer Patients[J]. Cancers (Basel), 2020, 12(4): 829. doi: 10.3390/cancers12040829
[31] Figlioli G, Kvist A, Tham E, et al. The Spectrum of FANCM Protein Truncating Variants in European Breast Cancer Cases[J]. Cancers (Basel), 2020, 12(2): 292. doi: 10.3390/cancers12020292
[32] Zhunussova G, Afonin G, Abdikerim S, et al. Mutation Spectrum of Cancer-Associated Genes in Patients With Early Onset of Colorectal Cancer[J]. Front Oncol, 2019, 9: 673. doi: 10.3389/fonc.2019.00673
[33] Xie Z, Ke Y, Chen J, et al. Prevalence and Spectrum of Predisposition Genes With Germline Mutations Among Chinese Patients With Bowel Cancer[J]. Front Genet, 2021, 12: 755629.
[34] Chakrabarti S, Kamgar M, Mahipal A. Systemic Therapy of Metastatic Pancreatic Adenocarcinoma: Current Status, Challenges, and Opportunities[J]. Cancers (Basel), 2022, 14(11): 2588. doi: 10.3390/cancers14112588
[35] Rasool R, Ullah I, Mubeen B, et al. Theranostic Interpolation of Genomic Instability in Breast Cancer[J]. Int J Mol Sci, 2022, 23(3): 1861. doi: 10.3390/ijms23031861
[36] Uson PLS, Jr, Kunze KL, Golafshar MA, et al. Germline Cancer Testing in Unselected Patients with Gastric and Esophageal Cancers: A Multi-center Prospective Study[J]. Dig Dis Sci, 2022, 67(11): 5107-5115. doi: 10.1007/s10620-022-07387-x
[37] Yang Z, Wu XS, Wei Y, et al. Transcriptional Silencing of ALDH2 Confers a Dependency on Fanconi Anemia Proteins in Acute Myeloid Leukemia[J]. Cancer Discov, 2021, 11(9): 2300-2315. doi: 10.1158/2159-8290.CD-20-1542
[38] Feng L, Jin F. Expression and prognostic significance of Fanconi anemia group D2 protein and breast cancer type 1 susceptibility protein in familial and sporadic breast cancer[J]. Oncol Lett, 2019, 17(4): 3687-3700.
[39] Li X, Liu J, Wang K, et al. Polymorphisms and rare variants identified by next-generation sequencing confer risk for lung cancer in han Chinese population[J]. Pathol Res Pract, 2020, 216(4): 152873. doi: 10.1016/j.prp.2020.152873
[40] Liu J, Mroczek M, Mach A, et al. Genetics, Genomics and Emerging Molecular Therapies of Pancreatic Cancer[J]. Cancers (Basel), 2023, 15(3): 779. doi: 10.3390/cancers15030779
[41] Alenezi WM, Fierheller CT, Serruya C, et al. Genetic analyses of DNA repair pathway associated genes implicate new candidate cancer predisposing genes in ancestrally defined ovarian cancer cases[J]. Front Oncol, 2023, 13: 1111191.
[42] Liu W, Palovcak A, Li F, et al. Fanconi anemia pathway as a prospective target for cancer intervention[J]. Cell Biosci, 2020, 10: 39.
[43] Miao H, Ren Q, Li H, et al. Comprehensive analysis of the autophagy-dependent ferroptosis-related gene FANCD2 in lung adenocarcinoma[J]. BMC Cancer, 2022, 22(1): 225.
[44] Zheng C, Ren Z, Chen H, et al. FANCD2 promotes the malignant behavior of endometrial cancer cells and its prognostic value[J]. Exp Cell Res, 2022, 421(2): 113388.
[45] Wu X, Liu G, Liu R, et al. Expression of ubiquitin-conjugating enzyme E2T in colorectal cancers and clinical implications[J]. Oncol Lett, 2020, 20(5): 275.
[46] Zhou Z, Yin H, Suye S, et al. Pan-cancer analysis of the prognostic and immunological role of Fanconi anemia complementation group E[J]. Front Genet, 2022, 13: 1024989.
[47] Gandin V, Hoeschele JD, Margiotta N. Special Issue "Cisplatin in Cancer Therapy: Molecular Mechanisms of Action 3.0"[J]. Int J Mol Sci, 2023, 24(9): 7917.
[48] Mangogna A, Munari G, Pepe F, et al. Homologous Recombination Deficiency in Ovarian Cancer: from the Biological Rationale to Current Diagnostic Approaches[J]. J Pers Med, 2023, 13(2): 284.
[49] Ali R, Aouida M, Alhaj Sulaiman A, et al. Can Cisplatin Therapy Be Improved? Pathways That Can Be Targeted[J]. Int J Mol Sci, 2022, 23(13): 7241.
[50] Huang Z, Chen Y, Chen R, et al. HPV Enhances HNSCC Chemosensitization by Inhibiting SERPINB3 Expression to Disrupt the Fanconi Anemia Pathway[J]. Adv Sci (Weinh), 2022, 10(1): e2202437.
[51] Yuan M, Wu Q, Zhang M, et al. Disulfiram enhances the antitumor activity of cisplatin by inhibiting the Fanconi anemia repair pathway[J]. J Zhejiang Univ Sci B, 2023, 24(3): 207-220.
[52] Zhang Y, Li J, Zhou Y, et al. And-1 Coordinates with the FANCM Complex to Regulate Fanconi Anemia Signaling and Cisplatin Resistance[J]. Cancer Res, 2022, 82(18): 3249-3262.
[53] Singh TD, Singh NI, Devi KM, et al. Planispine A Sensitized Cancer Cells to Cisplatin by Inhibiting the Fanconi Anemia Pathway[J]. Molecules, 2022, 27(21): 7288.
[54] Fan XZ, Chen YF, Zhang SB, et al. Centipeda minima extract sensitizes lung cancer cells to DNA-crosslinking agents via targeting Fanconi anemia pathway[J]. Phytomedicine, 2021, 91: 153689.
[55] Turner JG, Cui Y, Bauer AA, et al. Melphalan and Exportin 1 Inhibitors Exert Synergistic Antitumor Effects in Preclinical Models of Human Multiple Myeloma[J]. Cancer Res, 2020, 80(23): 5344-5354.



 下载:
下载:

