Effect of LncRNA LINC00857 Knockdown on Migration, Proliferation and Apoptosis of Pancreatic Cancer PANC-1 Cells
-
摘要:目的
探讨LncRNA LINC00857在胰腺细胞系中的表达及其下调对胰腺癌PANC-1细胞增殖、迁移和凋亡的影响,并探讨其机制。
方法构建慢病毒载体GV112,转染胰腺癌细胞株PANC-1为实验组,转染空白质粒为阴性对照组,未干预的细胞作为正常对照组;CCK-8法、Transwell实验、划痕实验、流式细胞术、Western blot检测下调LINC00857对细胞增殖、迁移、凋亡、细胞周期以及上皮间质转化(EMT)相关蛋白的影响。
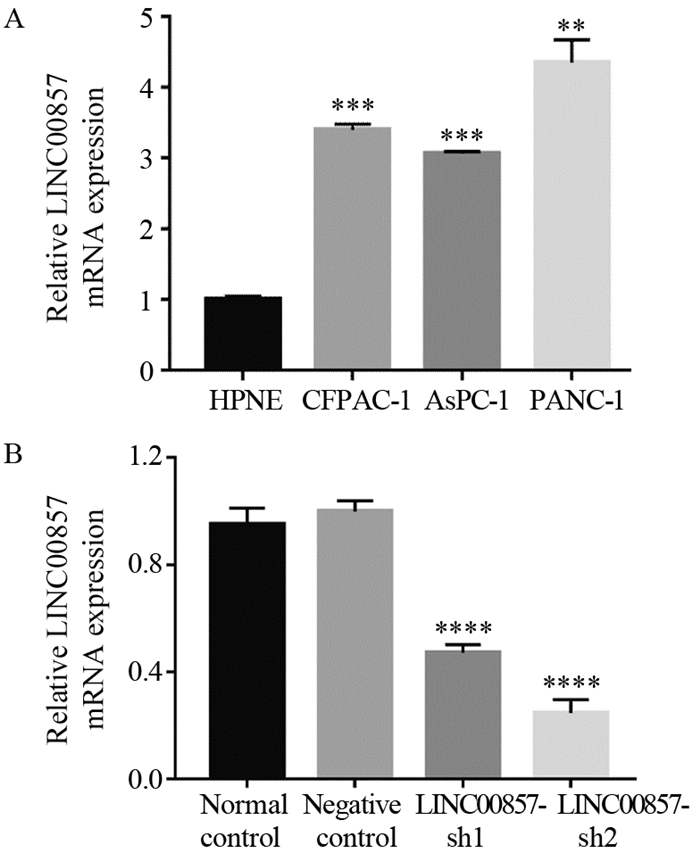
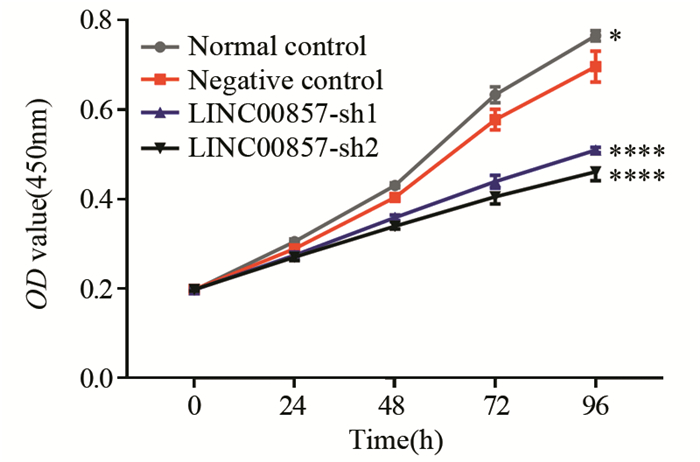
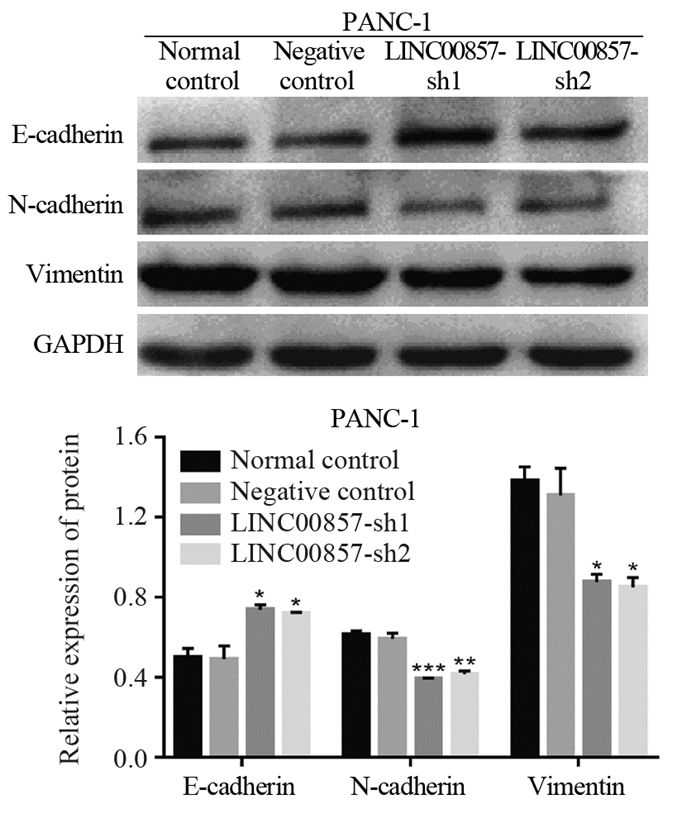
结果胰腺癌细胞系中LINC00857的表达显著高于胰腺正常上皮细胞(P < 0.01)。敲低LINC00857可显著抑制PANC-1细胞的增殖和迁移能力(P < 0.0001);与阴性对照组相比,实验组细胞的凋亡率明显增加(P < 0.05),G0/G1期细胞数量增多(P < 0.01),S期细胞数目减少(P < 0.05),细胞被阻滞在G1期;E-cadherin表达显著上调(P < 0.05),N-cadherin和Vimentin表达显著下调(N-cadherin P < 0.01, Vimentin P < 0.05)。
结论LINC00857可通过调控G1/S期转化EMT信号通路促进PANC-1细胞的增殖和迁移,并抑制其凋亡。
Abstract:ObjectiveTo investigate the expression of LncRNA LINC00857 in pancreatic cells and the effect of lncRNA LINC00857 down-regulation on proliferation, migration and apoptosis of pancreatic cancer PANC-1 cells and possible mechanism.
MethodsThe lentiviral vector GV112 was constructed and infected PANC-1 cells to obtain experimental group, while the blank plasmid was transfected as a negative control group and the cells without intervention were taken as normal control group. CCK-8 assay, Transwell assay, scratch test, flow cytometry, Western blot were used to detect the effect of LINC00857 down-regulation on cell proliferation, migration, apoptosis, cell cycle and EMT-related proteins.
ResultsLINC00857 expression in pancreatic cancer cells were significantly higher than that in normal pancreatic epithelial cells (P < 0.001). Knockdown of LINC00857 could significantly inhibit the proliferation and migration of PANC-1 cells (P < 0.0001); compared with the negative control group, the apoptosis rates of cells in the experimental groups were significantly increased (P < 0.05), the number of cells in G0/G1 phase increased (P < 0.01), the number of cells in S phase decreased (P < 0.05), and the cells were blocked in the G1 phase, E-cadherin expression was significantly up-regulated (P < 0.05) and N-cadherin and Vimentin expression were significantly down-regulated (N-cadherin: P < 0.01, Vimentin: P < 0.05).
ConclusionLINC00857 can promote proliferation and migration of PANC-1 cells and inhibit its apoptosis by regulating G1/S phase transition and EMT signaling pathway.
-
Key words:
- Pancreatic cancer /
- LncRNA LINC00857 /
- EMT /
- Apoptosis /
- Cell cycle /
- Proliferation /
- Migration
-
0 引言
胶质瘤是一种中枢神经系统肿瘤[1]。世界卫生组织(WHO) 将胶质瘤分为四个等级,其中Ⅳ级胶质瘤恶性程度最高,也称为胶质母细胞瘤(GBM),其两年生存率仅26%,中位生存期仅14.6个月[2-4]。探索胶质瘤进展的分子机制对于胶质瘤治疗具有重要意义。
各种转录因子可参与调控人类疾病的进展,目前可表达转录因子的基因约占所有致癌基因的20%[5-8]。E-twenty six(ETS)蛋白是转录因子家族中最大的一个家族之一,在调节多种生物学过程中发挥重要作用[9]。ETS原癌基因1(ETS proto-oncogene 1, ETS1)是RAS/RAF/ERK通路的下游效应子[10]。据报道ETS1可参与多种实体瘤的发生发展,如结直肠癌[11]、宫颈癌[12]、非小细胞肺癌[13]。在胶质瘤中,ETS1可通过激活长链非编码RNA(long non-coding RNA, lncRNA)SNHG10转录发挥促癌作用[14]。然而,ETS1在胶质瘤中的生物学功能和下游调控机制尚未完全阐明。
LncRNA XIST高表达可促进胶质瘤的恶性生物学表型[15]。本研究中,我们发现ETS1在胶质瘤组织和细胞内高表达,调控肿瘤细胞的增殖和凋亡,且能激活XIST的转录。这些发现揭示了ETS1新的生物学功能,并提示ETS1可能是胶质瘤的治疗靶点。
1 材料与方法
1.1 组织样本收集
收集2019年9月至2022年12月在三峡大学第一临床医学院神经外科收治的58例初诊胶质瘤手术患者的肿瘤组织及其相应的癌旁组织。所有组织样本取出后立即放入液氮中冷冻,随后转移至冰箱中(−80℃)保存。所有患者术前未接受放疗或化疗,患者或家属在组织样本收集前均签署了知情同意书。所有实验方案经本院伦理委员会批准,实验流程严格遵守赫尔辛基宣言。
1.2 细胞培养
胶质瘤细胞系(U251、U87、LN229、LN308)和人星形胶质细胞(NHA)均购自中国典型培养物保藏中心(武汉)。所有细胞系培养于含10%胎牛血清(美国西格玛-奥德里奇公司)、100 U/ml青霉素、100 μg/ml链霉素的培养基中,并在5%CO2、37℃的培养箱中孵育。取对数生长期的细胞用于实验。
1.3 细胞转染
ETS1小干扰RNA(si-ETS1#1和si-ETS1#2),XIST过表达质粒(XIST),ETS1过表达质粒(ETS1)和空白质粒(NC)均购自吉满生物科技有限公司(上海),当细胞融合达60%~80%时,使用LipofectamineTM2000(美国英杰公司)将以上siRNA或载体转染U87和LN229细胞系中,转染48 h后,采用实时定量聚合酶链反应(qRT-PCR)检测转染效率,收集细胞用于后续实验。
1.4 免疫组织化学
将石蜡包埋的组织标本切成4 μm厚的切片,二甲苯脱蜡,磷酸盐缓冲液(PBS)洗涤,在柠檬酸溶液中煮沸进行抗原修复,然后用3%过氧化氢处理。随后将组织标本与1%FBS孵育,加入抗ETS1抗体(1∶100),4℃下孵育过夜,随后加入辣根过氧化物酶标记的二抗,室温孵育30 min后在室温下用DAB孵育5 min进行显色,苏木精对比染色,光学显微镜下观察并拍照。
1.5 实时定量PCR
TRIzol试剂提取组织和细胞中的RNA,ABI High-Capacity cDNA反转录试剂盒对RNA进行反转录,ABI StepOnePlus实时PCR系统进行实时定量PCR,GAPDH作为内参, 2-ΔΔCt法计算结果。引物序列:ETS1(正向:5’-GATAGTTGTGATCGCCTCACC-3’; 反向:5’-GTCCTCTGAGTCGAAGCTGTC-3’);XIST(正向:5’-AGCTCCTCGGACAGCTGTAA-3’; 反向:5’-CTCCAGATAGCTGGCAACC-3’); GAPDH (正向:5’-TCGACAGTCAGCCGCATCTTCTTT-3’,反向:5’-ACCAAATCCGTTGACTCCGACCTT-3’)。
1.6 CCK-8实验
CCK-8试剂盒检测U87细胞系和LN229细胞活性,将细胞以密度为1×103细胞/孔接种于96孔板上,转染细胞培养24、48和72 h后每孔分别加入10 μl CCK-8试剂,之后在37℃、5%CO2条件下孵育2 h。使用酶标仪检测每孔在450 nm波长处的吸光度值。
1.7 5-乙炔基-2'-脱氧尿苷实验
EdU试剂盒检测U87和LN229细胞的增殖。取对数生长期细胞,以2×103个细胞接种于96孔板中,每孔加入200 μl浓度为5 μmol/L EdU溶液,培养2 h,PBS冲洗,随后4%多聚甲醛固定30 min,加入Apollo®荧光染色液,避光孵育30 min,DAPI反应液染色30 min。荧光显微镜下观察细胞,并计算EdU阳性细胞的百分比。
1.8 Western blot实验
RIPA裂解液从细胞中提取蛋白,二喹啉甲酸法(BCA法)定量蛋白。十二烷基硫酸钠聚丙烯酰胺凝胶电泳后,将蛋白转移至聚偏二氟乙烯薄膜上,之后将PVDF膜用5%脱脂牛奶封闭。然后,将膜与下列一抗于4℃条件下孵育过夜:anti-Bax(1:
1000 )、anti-Bak(1:1000 )、anti-Bcl-2(1:1000 )或anti-β-actin (1:1000 ),随后将膜与二抗(1:2000 )于室温孵育2 h后使用ECL化学发光检测试剂盒检测蛋白条带,β-actin为内参。1.9 双荧光素酶报告基因实验
将ETS1与XIST启动子区域上的结合位点克隆到萤火虫荧光素酶报告载体pGL3中,构建XIST野生型载体(XIST-WT)和XIST突变型载体(XIST-MUT)。随后根据制造商的说明,LipofectamineTM 2000将上述载体和ETS1过表达质粒(ETS1)或空白质粒(NC)共转染到U87和LN229细胞中。转染48 h后,使用双荧光素酶报告系统检测荧光素酶活性,以海肾荧光素酶活性为内参。
1.10 染色质免疫共沉淀-PCR
EZ-ChIPTM试剂盒进行ChIP实验。将U87和LN229细胞用1%甲醛固定10 min,加入甘氨酸终止固定。刮取细胞并离心以获得细胞沉淀,加入含PMSF的细胞裂解缓冲液悬浮细胞,离心后去掉上清液获得核沉淀物,冰浴超声裂解后,将裂解物分别与一抗 IgG(1∶100)或ETS1一抗在4℃下过夜。随后,蛋白质琼脂糖沉淀DNA-蛋白质复合物,并在4℃下离心5 min。最后,根据EZ-ChIPTM试剂盒操作说明提取和处理DNA,qRT-PCR检测XIST的DNA水平。
1.11 生物信息学分析
GEPIA(http://gepia.cancer-pku.cn/)分析ETS1在胶质瘤组织中的差异表达。UALCAN数据库(ualcan.path.uab.edu/home)分析ETS1在胶质瘤组织以及正常脑组织中的表达。PROMO数据库(http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo)预测ETS1与XIST启动子之间的结合位点。cBioPortal数据库(https://www.cbioportal.org/)分析ETS1与XIST在胶质瘤组织中表达的相关性。
1.12 统计学方法
采用SPSS 22.0软件进行统计分析。所有实验进行3次,结果以均数±标准差表示,组间比较使用Student's t test或单因素方差分析,Pearson进行相关性分析。P<0.05为差异有统计学意义。
2 结果
2.1 ETS1在胶质瘤中高表达
首先使用GEPIA数据库分析ETS1在胶质瘤中的差异表达,结果表明ETS1在胶质母细胞瘤(GBM)和低级别胶质瘤(LGG)组织中表达均显著上调,见图1A。另外,UALCAN数据库同样显示ETS1在GBM组织中表达上调,见图1B。IHC染色检测58例胶质瘤组织及其对照正常组织样本中ETS1的表达,结果表明与正常脑组织相比,ETS1在胶质瘤组织中的表达显著上调(χ2=8.838,P=0.003),见图1C。qRT-PCR分析ETS1 mRNA在58例胶质瘤患者肿瘤组织以及正常脑组织中的表达水平,结果显示ETS1 mRNA在肿瘤组织中的表达水平显著高于正常脑组织见图1D。根据胶质瘤组织中ETS1表达水平的中位数,将58例胶质瘤患者分为高表达组和低表达组,分析ETS1的表达与患者临床病理特征之间的关系,结果显示ETS1高表达与胶质瘤患者TNM分期增加有关,见表1。此外,与人星形胶质细胞(NHA)相比,ETS1 mRNA在4种胶质瘤细胞系(U251、 U87、 LN229、 LN308)中表达水平均显著升高 ,见图1E。
![]() 图 1 ETS1在胶质瘤中的表达特征Figure 1 Expression characteristics of ETS1 in glioma*: P<0.05, ***: P<0.001; A: the differential expression of ETS1 in glioblastoma and low-grade glioma was analyzed using GEPIA online database; B: the expression of ETS1 in glioma and its adjacent tissues was analyzed using UALCAN database; C: the expression of ETS1 in 58 glioma tissues and adjacent tissues was detected by immunohistochemistry; D-E: the ETS1 mRNA expression levels in 58 glioma tissues and cell lines were detected by qRT-PCR.表 1 ETS1在胶质瘤组织中的表达与临床病理资料的关系Table 1 Relationship between ETS1 expression in glioma tissues and clinicopathological data of patients
图 1 ETS1在胶质瘤中的表达特征Figure 1 Expression characteristics of ETS1 in glioma*: P<0.05, ***: P<0.001; A: the differential expression of ETS1 in glioblastoma and low-grade glioma was analyzed using GEPIA online database; B: the expression of ETS1 in glioma and its adjacent tissues was analyzed using UALCAN database; C: the expression of ETS1 in 58 glioma tissues and adjacent tissues was detected by immunohistochemistry; D-E: the ETS1 mRNA expression levels in 58 glioma tissues and cell lines were detected by qRT-PCR.表 1 ETS1在胶质瘤组织中的表达与临床病理资料的关系Table 1 Relationship between ETS1 expression in glioma tissues and clinicopathological data of patientsCharacteristics ETS1 χ2 P High expression
(n=29)Low expression
(n=29)Gender 0.633 0.426 Male 15 18 Female 14 11 Age(years) 1.105 0.293 ≥40 13 17 <40 16 12 Diameter of
tumors (cm)0.624 0.430 < 2 12 15 ≥2 17 14 No 6 17 WHO staging
system5.695 0.017 3-4 21 12 1-2 8 17 2.2 敲低ETS1抑制胶质瘤细胞增殖、促进细胞凋亡
为探究ETS1在胶质瘤细胞中的生物学功能,我们将两种ETS1 siRNA(si-ETS1#1、si-ETS1#2)转染入U87和LN229细胞系中,qRT-PCR结果显示转染成功,见图2A。CCK-8实验显示,与si-NC组相比,si-ETS1#1和si-ETS1#2组中胶质瘤细胞的活性显著降低,见图2B。EdU实验同样显示si-ETS1#1和si-ETS1#2组中胶质瘤细胞增殖能力显著低于si-NC组,见图2C。Western blot实验结果显示,与si-NC组相比,si-ETS1#1和si-ETS1#2组细胞中促凋亡蛋白Bax和Bak的表达水平显著升高,而抗凋亡蛋白Bcl-2的表达显著降低,见图2D。因此,以上结果显示ETS1可促进胶质瘤细胞增殖,并抑制凋亡。
![]() 图 2 敲低ETS1抑制胶质瘤细胞增殖、促进细胞凋亡Figure 2 ETS1 knockdown inhibited glioma cell proliferation and promoted cell apoptosis*: P<0.05; **: P<0.01; ***: P<0.001; A: qRT-PCR was used to detect the expression level of ETS1 mRNA in U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; B: CCK-8 assay was conducted to detect the activity of U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; C: EdU assay was used to detect the proliferation of U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; D: Western blot was adopted to detect the expression levels of apoptosis-related proteins (Bax, Bak, Bcl-2) in U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2.
图 2 敲低ETS1抑制胶质瘤细胞增殖、促进细胞凋亡Figure 2 ETS1 knockdown inhibited glioma cell proliferation and promoted cell apoptosis*: P<0.05; **: P<0.01; ***: P<0.001; A: qRT-PCR was used to detect the expression level of ETS1 mRNA in U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; B: CCK-8 assay was conducted to detect the activity of U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; C: EdU assay was used to detect the proliferation of U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; D: Western blot was adopted to detect the expression levels of apoptosis-related proteins (Bax, Bak, Bcl-2) in U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2.2.3 ETS1靶向结合XIST激活其转录
为探索ETS1的下游机制,我们检索了PROMO数据库,发现ETS1可能与XIST启动子序列结合,见图3A;cBioPortal数据库结果显示,在胶质瘤组织中XIST与ETS1 mRNA的表达呈正相关,见图3B。通过双荧光素酶报告基因实验验证ETS1与XIST的结合关系,结果显示在U87和LN229细胞系中,过表达ETS1均可显著提高XIST WT报告质粒的荧光素酶活性,见图3C。在U87和LN229细胞系中使用ETS1特异性抗体进行ChIP-qPCR检测进一步证实二者的结合关系,结果表明与IgG对照组相比,ETS1抗体可以显著富集XIST的启动子序列,见图3D。将si-ETS1#1、si-ETS1#2转染进U87和LN229细胞系中,qRT-PCR结果显示与对照组相比,敲低ETS1可显著降低XIST的表达,见图3E。此外,qRT-PCR结果显示,XIST在胶质瘤组织中高表达,见图3F;且Pearson分析发现在胶质瘤组织中XIST与ETS1 mRNA的表达呈正相关,见图3G;qRT-PCR结果显示,与正常细胞相比,XIST在胶质瘤细胞中表达上调,见图3H。这些数据表明,ETS1可靶向结合XIST促进其表达。
![]() 图 3 ETS1靶向结合XISTFigure 3 ETS1 targeted XIST**: P<0.01; ***: P<0.001; A: PROMO database predicted the binding site of ETS1 to the XIST promoter; B: cBioPortal database was used to analyze the correlation between ETS1 mRNA and XIST expression in glioma tissues; C: dual-luciferase reporter assay was conducted to examine the effect of ETS1 overexpression on the luciferase activity of XIST-WT and XIST-MUT; D: ChIP-qPCR was used to detect the binding of ETS1 to the XIST promoter region; E: qRT-PCR was utilized to detect the expression level of XIST in U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; F: qRT-PCR was used to detect the expression level of XIST in 58 glioma tissues and adjacent tissues; G: Pearson correlation analysis was performed on the expression of ETS1 mRNA and XIST in glioma tissues; H: the expression level of XIST in glioma cell lines and NHA cells was detected by qRT-PCR.
图 3 ETS1靶向结合XISTFigure 3 ETS1 targeted XIST**: P<0.01; ***: P<0.001; A: PROMO database predicted the binding site of ETS1 to the XIST promoter; B: cBioPortal database was used to analyze the correlation between ETS1 mRNA and XIST expression in glioma tissues; C: dual-luciferase reporter assay was conducted to examine the effect of ETS1 overexpression on the luciferase activity of XIST-WT and XIST-MUT; D: ChIP-qPCR was used to detect the binding of ETS1 to the XIST promoter region; E: qRT-PCR was utilized to detect the expression level of XIST in U87 and LN229 cells after transfection with si-ETS1#1 or si-ETS1#2; F: qRT-PCR was used to detect the expression level of XIST in 58 glioma tissues and adjacent tissues; G: Pearson correlation analysis was performed on the expression of ETS1 mRNA and XIST in glioma tissues; H: the expression level of XIST in glioma cell lines and NHA cells was detected by qRT-PCR.2.4 过表达XIST可逆转敲低ETS1对胶质瘤细胞增殖和凋亡的影响
为进一步证实ETS1是否通过调控XIST促进胶质瘤细胞的增殖,我们将si-ETS1#1和XIST过表达质粒共转染U87和LN229细胞系,并通过qRT-PCR验证转染效率,见图4A。转染成功后,CCK-8和EdU实验结果显示,与单独转染si-ETS1#1组相比,共转染si-ETS1#1和XIST组中胶质瘤细胞增殖能力显著提高,见图4B、C。Western blot实验结果显示,与单独转染si-ETS1#1组相比,共转染si-ETS1#1和XIST组细胞中Bax、Bak的表达水平显著降低,而Bcl-2的表达显著升高,见图4D。结果表明过表达XIST可部分逆转敲低ETS1对胶质瘤细胞增殖的抑制作用,以及对细胞凋亡的促进作用,EST1可以依赖XIST调控胶质瘤细胞的增殖和凋亡。
![]() 图 4 过表达XIST可逆转敲低ETS1对胶质瘤细胞增殖和凋亡的影响Figure 4 Overexpression of XIST reversed the effect of ETS1 knockdown on the proliferation and apoptosis of glioma cells*: P<0.05; **: P<0.01; ***: P<0.001; A: qRT-PCR was used to detect the expression level of XIST in U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST; B: CCK-8 assay was performed to detect the activity of U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST; C: EdU assay was used to detect the proliferation of U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST; D: Western blot was employed to detect the expression levels of apoptosis-related proteins in U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST.
图 4 过表达XIST可逆转敲低ETS1对胶质瘤细胞增殖和凋亡的影响Figure 4 Overexpression of XIST reversed the effect of ETS1 knockdown on the proliferation and apoptosis of glioma cells*: P<0.05; **: P<0.01; ***: P<0.001; A: qRT-PCR was used to detect the expression level of XIST in U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST; B: CCK-8 assay was performed to detect the activity of U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST; C: EdU assay was used to detect the proliferation of U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST; D: Western blot was employed to detect the expression levels of apoptosis-related proteins in U87 and LN229 cells after co-transfection of si-ETS1#1 and XIST.3 讨论
转录因子是一种DNA结合蛋白,通过结合特定区域来调节基因的表达[16]。长期以来,由于转录因子主要定位于细胞质和细胞核,除了配体诱导的核受体外,转录因子曾被认为是“不可成药”的靶点[17]。近年,很多研究发现特异性小分子可以阻断转录因子和DNA、蛋白质的相互作用,有些化合物可以诱导转录因子通过泛素化降解,这些发现为靶向转录因子治疗肿瘤开辟了新的可能性[18]。
最近的几项研究表明转录因子ETS1可参与胶质瘤的进展。如ETS1通过结合在SNHG10启动子区域促进其转录,从而上调FBXL19的表达,最终促进胶质瘤细胞的增殖、迁移和侵袭[14];PAXIP1-AS1通过募集ETS1上调KIF14的表达,从而促进胶质瘤细胞的迁移和侵袭以及血管生成[19]。ETS1能作为果糖1,6二磷酸酶基因的转录抑制因子抑制其表达,介导糖酵解和肿瘤细胞侵袭迁移[20]。还有研究发现ETS1在胶质母细胞瘤衍生的内皮和间充质干细胞样细胞中高表达,且其表达与胶质母细胞瘤的侵袭性以及微血管密度相关[21]。本研究中,我们发现ETS1在胶质瘤组织和细胞系中高表达,且其高表达与胶质瘤患者不良临床病理指标有关。另外,敲低ETS1可显著抑制胶质瘤细胞的增殖,并促进细胞凋亡。我们的研究结果与既往的报道[14,19-21]均提示,ETS1可能成为胶质瘤的潜在治疗靶点。
本研究证实了ETS1可与XIST的启动子结合。lncRNA是一种长度超过200个核苷酸的非编码RNA,大量研究表明lncRNAs可参与多种疾病的发生发展[22-24]。XIST是lncRNA之一,研究发现其可调控肿瘤细胞的恶性表型[25]。XIST已被证实在许多肿瘤中上调,如在结直肠癌中,XIST充当致癌因子,通过抑制miR-132-3p的表达促进癌细胞的增殖[26];甲状腺癌中,XIST在癌组织和细胞系中均显著上调,并通过调控miR-34a和MET/PI3K/AKT信号通路促进肿瘤细胞增殖[27];非小细胞肺癌中,XIST在患者组织样本中高表达,敲低XIST可抑制癌细胞的生长,并促进肿瘤细胞对顺铂的敏感度[28]。研究表明XIST在胶质瘤中上调,且通过miR-133a/SOX4轴促进细胞的增殖和转移[15];还有研究发现过表达XIST可通过调控miR-329/CREB1轴促进胶质瘤细胞增殖、侵袭,并抑制细胞凋亡及降低癌细胞的放射敏感度[29];敲低XIST可通过上调miR-204-5p的表达,进而抑制胶质瘤细胞的增殖、迁移和侵袭,并促进细胞凋亡,从而抑制胶质瘤的进展[30]。本研究中,同样发现XIST在胶质瘤组织和细胞中显著上调,且生物信息学分析显示在胶质瘤组织中ETS1与XIST的表达呈正相关,且XIST的转录受ETS1调控,过表达XIST可部分逆转敲低ETS1对胶质瘤细胞恶性表型的抑制作用。本研究为XIST在胶质瘤中的异常表达提供了一种合理的解释。
总之,本研究证实ETS1在胶质瘤组织中高表达并与患者不良临床病理指标相关,过表达ETS1可以促进XIST的转录,这表明ETS1在胶质瘤进展中发挥潜在促癌作用。
Competing interests: The authors declare that they have no competing interests.作者贡献:张波:设计与实施实验、收集与分析数据、撰写论文任龙飞:指导数据分析和论文写作胡进静、白仲添:指导实验周文策:指导实验设计、修改和审核论文 -
-
[1] Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019[J]. CA Cancer J Clin, 2019, 69(1): 7-34. doi: 10.3322/caac.21551
[2] Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68(6): 394-424. doi: 10.3322/caac.21492
[3] Zhou W, Chen L, Li C, et al. The multifaceted roles of long noncoding RNAs in pancreatic cancer: an update on what we know[J]. Cancer Cell Int, 2020, 20: 41. doi: 10.1186/s12935-020-1126-1
[4] Wang L, He Y, Liu W, et al. Non-coding RNA LINC00857 is predictive of poor patientsurvival and promotes tumor progression via cell cycle regulation in lung cancer[J]. Oncotarget, 2016, 7(10): 11487-11499. doi: 10.18632/oncotarget.7203
[5] Wang L, Cao L, Wen C, et al. LncRNA LINC00857 regulates lung adenocarcinoma progression, apoptosis and glycolysis by targeting miR-1179/SPAG5 axis[J]. Hum Cell, 2020, 33(1): 195-204. doi: 10.1007/s13577-019-00296-8
[6] Zhang K, Shi H, Xi H, et al. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer[J]. Theranostics, 2017, 7(1): 213-227. doi: 10.7150/thno.16044
[7] Pang K, Ran MJ, Zou FW, et al. Long non-coding RNA LINC00857 promotes gastric cancer cell proliferation and predicts poor patient survival[J]. Oncol Lett, 2018, 16(2): 2119-2124.
[8] Dudek AM, van Kampen JGM, Witjes JA, et al. LINC00857 expression predicts and mediates the response to platinum-based chemotherapy in muscle-invasive bladder cancer[J]. Cancer Med, 2018, 7(7): 3342-3350. doi: 10.1002/cam4.1570
[9] Xia C, Zhang XY, Liu W, et al. LINC00857 contributes to hepatocellular carcinoma malignancy via enhancing epithelial-mesenchymal transition[J]. J Cell Biochem, 2018. Online ahead of print.
[10] Su W, Wang L, Niu F, et al. LINC00857 knockdown inhibits cell proliferation and induces apoptosis via involving STAT3 and MET oncogenic proteins in esophageal adenocarcinoma[J]. Aging (Albany NY), 2019, 11(9): 2812-2821.
[11] Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses[J]. Nucleic Acids Res, 2017, 45(W1): W98-W102. doi: 10.1093/nar/gkx247
[12] Li X, Zhou S, Fan T, et al. lncRNA DGCR 5/miR-27a-3p/BNIP3 promotes cell apoptosis in pancreatic cancer by regulating the p38 MAPK pathway[J]. Int J Mol Med, 2020, 46(2): 729-739. doi: 10.3892/ijmm.2020.4632
[13] Wang F, Rong L, Zhang Z, et al. LncRNA H19-Derived miR-675-3p Promotes Epithelial-Mesenchymal Transition and Stemness in Human Pancreatic Cancer Cells by targeting the STAT3 Pathway[J]. J Cancer, 2020, 11(16): 4771-4782. doi: 10.7150/jca.44833
[14] Vaidya AM, Sun Z, Ayat N, et al. Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy[J]. Bioconjug Chem, 2019, 30(3): 907-919. doi: 10.1021/acs.bioconjchem.9b00028
[15] 张志鹏, 孙维佳, 陈泓西, 等. 基于生物信息学的胰腺导管腺癌预后风险长链非编码RNA筛选[J]. 中国普通外科杂志, 2018, 27(9): 1126-1134. https://www.cnki.com.cn/Article/CJFDTOTAL-ZPWZ201809010.htm Zhang ZP, Sun WJ, Chen HX, et al. Screening of long non-coding RNA for prognostic risk of pancreatic ductal adenocarcinoma based on bioinformatics[J]. Zhongguo Pu Tong Wai Ke Za Zhi, 2018, 27(9): 1126-1134. https://www.cnki.com.cn/Article/CJFDTOTAL-ZPWZ201809010.htm
[16] Expósito-Villén A, Aránega AE, Franco D. Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition[J]. Noncoding RNA, 2018, 4(2): 1-14.
[17] 成鉴晓, 江月萍, 任琳琳, 等. lncRNA MEG3通过调控miR-543/PTEN分子轴抑制胰腺癌细胞增殖及转移的机制[J]. 肿瘤防治研究, 2019, 46(7): 588-593. doi: 10.3971/j.issn.1000-8578.2019.18.1876 Cheng JX, Jiang YP, Ren LL, et al. LncRNA MEG3 suppresses proliferation and metastasis of pancreatic cancer cells via regulating miR-543/PTEN axis[J]. Zhong Liu Fang Zhi Yan Jiu, 2019, 46(7): 588-593. doi: 10.3971/j.issn.1000-8578.2019.18.1876
[18] Cao W, Zhou G. LncRNA SNHG12 contributes proliferation, invasion and epithelial-mesenchymal transition of pancreatic cancer cells by absorbing miRNA-320b[J]. Biosci Rep, 2020, 40(6): BSR20200805. doi: 10.1042/BSR20200805




 下载:
下载: